6K4N
 
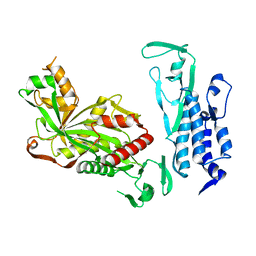 | | Cryo-EM structure of p300 | | Descriptor: | Histone acetyltransferase p300 | | Authors: | Ghosh, R, Roy, S, Sengupta, J. | | Deposit date: | 2019-05-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Tumor Suppressor p53-Mediated Structural Reorganization of the Transcriptional Coactivator p300.
Biochemistry, 58, 2019
|
|
3BCN
 
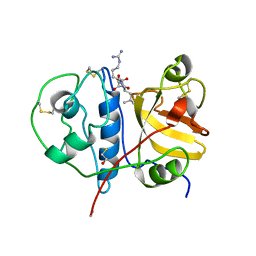 | | Crystal structure of a papain-like cysteine protease Ervatamin-A complexed with irreversible inhibitor E-64 | | Descriptor: | BETA-MERCAPTOETHANOL, Ervatamin-A, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Ghosh, R, Chakrabarti, C, Dattagupta, J.K, Biswas, S. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria
Febs J., 275, 2008
|
|
5XZC
 
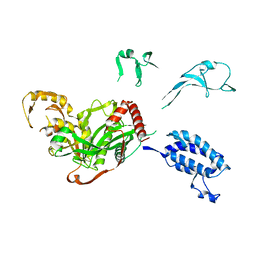 | | Cryo-EM structure of p300-p53 protein complex | | Descriptor: | Cellular tumor antigen p53, Histone acetyltransferase p300 | | Authors: | Ghosh, R, Roy, S, Sengupta, J. | | Deposit date: | 2017-07-12 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Tumor suppressor p53-mediated structural reorganization of the transcriptional coactivator p300.
Biochemistry, 2019
|
|
2PNS
 
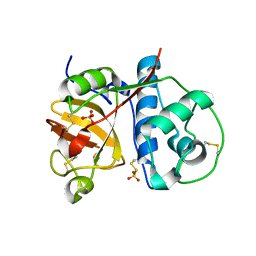 | | 1.9 Angstrom resolution crystal structure of a plant cysteine protease Ervatamin-C refinement with cDNA derived amino acid sequence | | Descriptor: | Ervatamin-C, a papain-like plant cysteine protease, PHOSPHATE ION, ... | | Authors: | Ghosh, R, Guha Thakurta, P, Biswas, S, Chakrabarti, C, Dattagupta, J.K. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A thermostable cysteine protease precursor from a tropical plant contains an unusual C-terminal propeptide: cDNA cloning, sequence comparison and molecular modeling studies.
Biochem.Biophys.Res.Commun., 362, 2007
|
|
2PRE
 
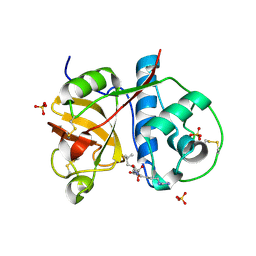 | | Crystal structure of plant cysteine protease Ervatamin-C complexed with irreversible inhibitor E-64 at 2.7 A resolution | | Descriptor: | Ervatamin-C, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, SULFATE ION | | Authors: | Ghosh, R, Chakrabarti, C, Dattagupta, J.K, Biswas, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate specificity and activity of ervatamins, the papain-like cysteine proteases from a tropical plant, Ervatamia coronaria.
Febs J., 275, 2008
|
|
2I0W
 
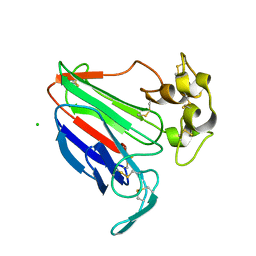 | |
7BMK
 
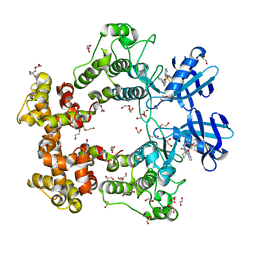 | | ATP-Competitive Partial Antagonists-'PAIR's-Rheostatically Modulate IRE1alpha's Kinase Helix-alphaC to Segregate its RNase-Mediated Biological Outputs | | Descriptor: | 1,2-ETHANEDIOL, 2,2,2-tris(fluoranyl)-~{N}-[4-[3-[2-[[(3~{S})-piperidin-3-yl]amino]pyrimidin-4-yl]pyridin-2-yl]oxynaphthalen-1-yl]ethanesulfonamide, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feldman, H.C, Ghosh, R, Auyeung, V, Mueller, J.L, Vidadala, V.N, Olivier, A, Backes, B.J, Zikherman, J, Papa, F.R, Maly, D.J. | | Deposit date: | 2021-01-20 | | Release date: | 2021-09-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | ATP-competitive partial antagonists of the IRE1 alpha RNase segregate outputs of the UPR.
Nat.Chem.Biol., 17, 2021
|
|
2HRI
 
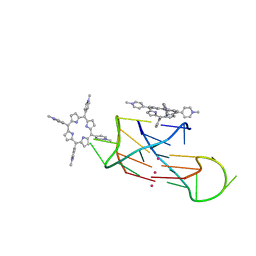 | | A parallel stranded human telomeric quadruplex in complex with the porphyrin TMPyP4 | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3', POTASSIUM ION | | Authors: | Parkinson, G.N, Ghosh, R, Neidle, S. | | Deposit date: | 2006-07-20 | | Release date: | 2007-05-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural basis for binding of porphyrin to human telomeres.
Biochemistry, 46, 2007
|
|
1ORD
 
 | | CRYSTALLOGRAPHIC STRUCTURE OF A PLP-DEPENDENT ORNITHINE DECARBOXYLASE FROM LACTOBACILLUS 30A TO 3.1 ANGSTROMS RESOLUTION | | Descriptor: | ORNITHINE DECARBOXYLASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hackert, M.L, Momany, C, Ernst, S, Ghosh, R. | | Deposit date: | 1995-02-08 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystallographic structure of a PLP-dependent ornithine decarboxylase from Lactobacillus 30a to 3.0 A resolution.
J.Mol.Biol., 252, 1995
|
|
8URK
 
 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrates | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA1, DNA2, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2023-10-26 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
9C3U
 
 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrate containing GTTTAC as recognition sequence | | Descriptor: | DNA1, DNA2, Methyltransferase, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2024-06-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
9C3T
 
 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrate containing GTAAAC as recognition sequence | | Descriptor: | DNA1, DNA2, Methyltransferase, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2024-06-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
9C3S
 
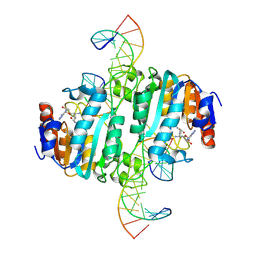 | | Crystal structure of DNA N6-Adenine Methyltransferase M.BceJIV from Burkholderia cenocepacia in complex with duplex DNA substrate containing GTATAC as recognition sequence | | Descriptor: | DNA1, DNA2, Methyltransferase, ... | | Authors: | Kottur, J, Quintana-Feliciano, R, Aggarwal, A.K. | | Deposit date: | 2024-06-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Burkholderia cenocepacia epigenetic regulator M.BceJIV simultaneously engages two DNA recognition sequences for methylation.
Nat Commun, 15, 2024
|
|
1MPF
 
 | |
5D1O
 
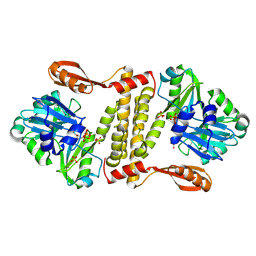 | | Archaeal ATP-dependent RNA ligase - form 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA ligase, MAGNESIUM ION, ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
5D1P
 
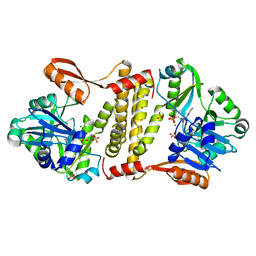 | | Archaeal ATP-dependent RNA ligase - form 2 | | Descriptor: | ATP-dependent RNA ligase, MAGNESIUM ION, SULFATE ION | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
1BT9
 
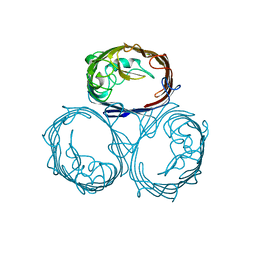 | | OMPF PORIN MUTANT D74A | | Descriptor: | PROTEIN (MATRIX PORIN OUTER MEMBRANE PROTEIN F) | | Authors: | Philippsen, A, Schirmer, T. | | Deposit date: | 1998-09-01 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Stability of trimeric OmpF porin: the contributions of the latching loop L2.
Biochemistry, 37, 1998
|
|
3EM2
 
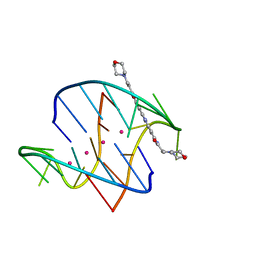 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6038 | | Descriptor: | 3,6-Bis[(3-morpholinopropionamido)] acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-09-23 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EQW
 
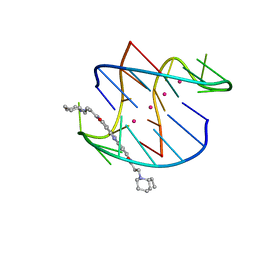 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in small unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-01 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ET8
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6054 | | Descriptor: | 3,6-Bis{3-(3-[(3R)-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EUM
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6066 | | Descriptor: | 3,6-Bis[3-(azepan-1-yl)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3EUI
 
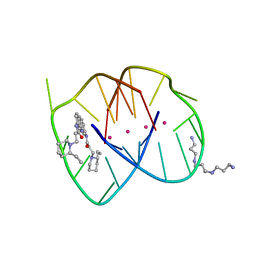 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6042 in a large unit cell | | Descriptor: | 3,6-Bis{3-[(2R)-(2-ethylpiperidino)]propionamido}acridine, 3-[(2R)-2-ethylpiperidin-1-yl]-N-[6-({3-[(2S)-2-ethylpiperidin-1-yl]propanoyl}amino)acridin-3-yl]propanamide, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', ... | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-10 | | Release date: | 2008-12-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ERU
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6045 | | Descriptor: | 3,6-Bis{3-[(2R)-2-methylpiperidino)]propionamido}acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
3ES0
 
 | | A bimolecular anti-parallel-stranded Oxytricha nova telomeric quadruplex in complex with a 3,6-disubstituted acridine BSU-6048 | | Descriptor: | 3,6-Bis[3-(4-methylpiperidino)propionamido]acridine, 5'-D(*DGP*DGP*DGP*DGP*DTP*DTP*DTP*DTP*DGP*DGP*DGP*DG)-3', POTASSIUM ION | | Authors: | Campbell, N.H, Parkinson, G, Neidle, S. | | Deposit date: | 2008-10-03 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selectivity in Ligand Recognition of G-Quadruplex Loops.
Biochemistry, 48, 2009
|
|
1PHO
 
 | |
