5A5T
 
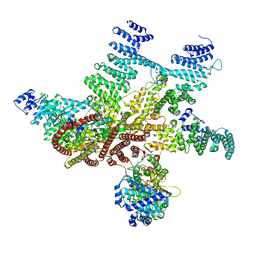 | | Structure of mammalian eIF3 in the context of the 43S preinitiation complex | | Descriptor: | EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT A, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT C, EUKARYOTIC TRANSLATION INITIATION FACTOR 3 SUBUNIT E, ... | | Authors: | des-Georges, A, Dhote, V, Kuhn, L, Hellen, C.U.T, Pestova, T.V, Frank, J, Hashem, Y. | | Deposit date: | 2015-06-21 | | Release date: | 2015-09-09 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of Mammalian Eif3 in the Context of the 43S Preinitiation Complex.
Nature, 525, 2015
|
|
4V4B
 
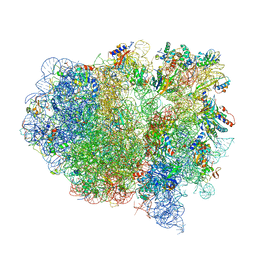 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
6WEJ
 
 | | Structure of cGMP-unbound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEK
 
 | | Structure of cGMP-bound WT TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CYCLIC GUANOSINE MONOPHOSPHATE, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6WEL
 
 | | Structure of cGMP-unbound F403V/V407A mutant TAX-4 reconstituted in lipid nanodiscs | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cyclic nucleotide-gated cation channel, ... | | Authors: | Zheng, X, Fu, Z, Su, D, Zhang, Y, Li, M, Pan, Y, Li, H, Li, S, Grassucci, R.A, Ren, Z, Hu, Z, Li, X, Zhou, M, Li, G, Frank, J, Yang, J. | | Deposit date: | 2020-04-02 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of ligand activation of a eukaryotic cyclic nucleotide-gated channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G34
 
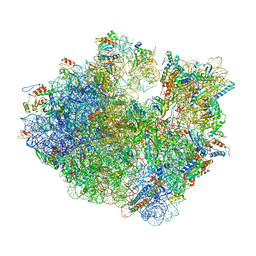 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:1st intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G31
 
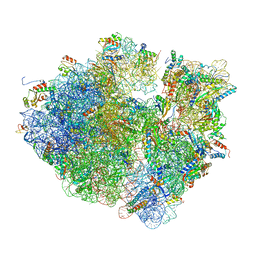 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:2nd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8G38
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:3rd Intermediate | | Descriptor: | 16S, 23S, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-07 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
3DNY
 
 | |
4V6Q
 
 | | Structural characterization of mRNA-tRNA translocation intermediates (class 5 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1JQT
 
 | | Fitting of L11 protein in the low resolution cryo-EM map of E.coli 70S ribosome | | Descriptor: | 50S Ribosomal protein L11 | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
3DG2
 
 | |
3DG4
 
 | |
3DG0
 
 | |
3DG5
 
 | |
6RS4
 
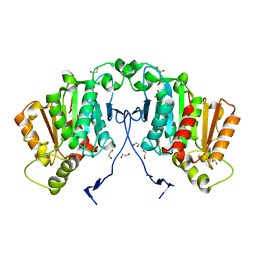 | | Structure of tabersonine synthase - an alpha-beta hydrolase from Catharanthus roseus | | Descriptor: | 1,2-ETHANEDIOL, Tabersonine synthase | | Authors: | Caputi, L, Franke, J, Bussey, K, Farrow, S.C, Curcino Vieira, I.J, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-05-21 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
6RT8
 
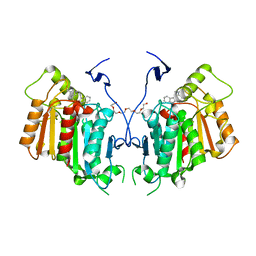 | | Structure of catharanthine synthase - an alpha-beta hydrolase from Catharanthus roseus with a cleaviminium intermediate bound | | Descriptor: | 18-carboxymethoxy-cleaviminium, Catharanthine synthase, HEXAETHYLENE GLYCOL | | Authors: | Caputi, L, Franke, J, Bussey, K, Farrow, S.C, Curcino Vieira, I.J, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-05-22 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
7SIN
 
 | | Structure of negative allosteric modulator-bound inactive human calcium-sensing receptor | | Descriptor: | 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, Isoform 1 of Extracellular calcium-sensing receptor | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SIL
 
 | | Structure of positive allosteric modulator-bound active human calcium-sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-chlorophenyl)-N-[(1R)-1-(3-methoxyphenyl)ethyl]propan-1-amine, ... | | Authors: | Park, J, Zuo, H, Frangaj, A, Fu, Z, Yen, L.Y, Zhang, Z, Mosyak, L, Slavkovich, V.N, Liu, J, Ray, K.M, Cao, B, Vallese, F, Geng, Y, Chen, S, Grassucci, R, Dandey, V.P, Tan, Y.Z, Eng, E, Lee, Y, Kloss, B, Liu, Z, Hendrickson, W.A, Potter, C.S, Carragher, B, Graziano, J, Conigrave, A.D, Frank, J, Clarke, O.B, Fan, Q.R. | | Deposit date: | 2021-10-14 | | Release date: | 2022-01-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Symmetric activation and modulation of the human calcium-sensing receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1ZN1
 
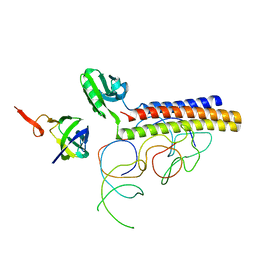 | | Coordinates of RRF fitted into Cryo-EM map of the 70S post-termination complex | | Descriptor: | 30S ribosomal protein S12, Ribosome recycling factor, ribosomal 16S RNA, ... | | Authors: | Gao, N, Zavialov, A.V, Li, W, Sengupta, J, Valle, M, Gursky, R.P, Ehrenberg, M, Frank, J. | | Deposit date: | 2005-05-11 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies.
Mol.Cell, 18, 2005
|
|
1JQM
 
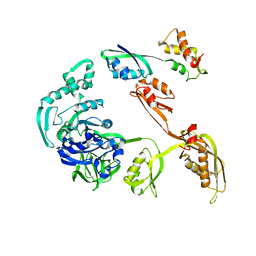 | | Fitting of L11 protein and elongation factor G (EF-G) in the cryo-em map of e. coli 70S ribosome bound with EF-G, GDP and fusidic acid | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
1JQS
 
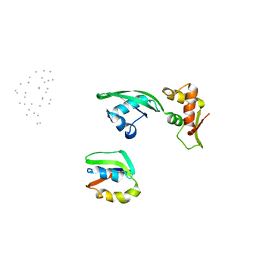 | | Fitting of L11 protein and elongation factor G (domain G' and V) in the cryo-em map of E. coli 70S ribosome bound with EF-G and GMPPCP, a nonhydrolysable GTP analog | | Descriptor: | 50S Ribosomal protein L11, Elongation Factor G | | Authors: | Agrawal, R.K, Linde, J, Segupta, J, Nierhaus, K.H, Frank, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-09-07 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation.
J.Mol.Biol., 311, 2001
|
|
6XE0
 
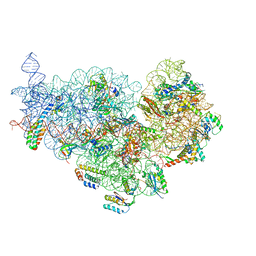 | | Cryo-EM structure of NusG-CTD bound to 70S ribosome (30S: NusG-CTD fragment) | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Washburn, R, Zuber, P, Sun, M, Hashem, Y, Shen, B, Li, W, Harvey, S, Acosta-Reyes, F.J, Knauer, S.H, Frank, J, Gottesman, M.E. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Escherichia coli NusG Links the Lead Ribosome with the Transcription Elongation Complex.
Iscience, 23, 2020
|
|
7SYP
 
 | | Structure of the wt IRES and 40S ribosome binary complex, open conformation. Structure 10(wt) | | Descriptor: | 18S rRNA, HCV IRES, HCV IRES partially loaded mRNA portion, ... | | Authors: | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | Deposit date: | 2021-11-25 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
