8SDD
 
 | | Crystal structure of fluoroacetate dehalogenase Daro3835 H274N mutant with D107-glycolyl intermediate | | 分子名称: | Alpha/beta hydrolase fold protein | | 著者 | Stogios, P.J, Skarina, T, Khusnutdinova, A, Iakounine, A, Savchenko, A. | | 登録日 | 2023-04-06 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural insights into hydrolytic defluorination of difluoroacetate by microbial fluoroacetate dehalogenases.
Febs J., 290, 2023
|
|
2P5X
 
 | | Crystal structure of Maf domain of human N-acetylserotonin O-methyltransferase-like protein | | 分子名称: | N-acetylserotonin O-methyltransferase-like protein, PHOSPHATE ION | | 著者 | Min, J, Wu, H, Dombrovski, L, Loppnau, P, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | 登録日 | 2007-03-16 | | 公開日 | 2007-03-27 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Biochemical and structural studies of conserved maf proteins revealed nucleotide pyrophosphatases with a preference for modified nucleotides.
Chem.Biol., 20, 2013
|
|
6D33
 
 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyribose-phosphate aldolase, GLYCEROL | | 著者 | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | 登録日 | 2018-04-14 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
2R6O
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | 著者 | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2007-09-06 | | 公開日 | 2007-09-18 | | 最終更新日 | 2012-10-24 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
7S2I
 
 | | Crystal structure of sulfonamide resistance enzyme Sul1 in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Kim, Y, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2K
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 in complex with 7,8-dihydropteroate, magnesium, and pyrophosphate | | 分子名称: | 4-AMINOBENZOIC ACID, 7,8-DIHYDROPTEROATE, CHLORIDE ION, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2M
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, Sul3 | | 著者 | Stogios, P.J, Skarina, T, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2L
 
 | | Crystal structure of sulfonamide resistance enzyme Sul3 apoenzyme | | 分子名称: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | 著者 | Stogios, P.J, Venkatesan, M, Michalska, K, Mesa, N, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7S2J
 
 | | Crystal structure of sulfonamide resistance enzyme Sul2 apoenzyme | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Michalska, K, Venkatesan, M, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-09-03 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
7TQ1
 
 | | Crystal structure of adaptive laboratory evolved sulfonamide-resistant Dihydropteroate Synthase (DHPS) from Escherichia coli in complex with 6-hydroxymethylpterin | | 分子名称: | 6-HYDROXYMETHYLPTERIN, Dihydropteroate synthase | | 著者 | Stogios, P.J, Skarina, T, Tan, K, Venkatesan, M, Fruci, M, Joachimiak, A, Savchenko, A, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-26 | | 公開日 | 2023-05-03 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | Molecular mechanism of plasmid-borne resistance to sulfonamide antibiotics.
Nat Commun, 14, 2023
|
|
3QVM
 
 | | The structure of olei00960, a hydrolase from Oleispira antarctica | | 分子名称: | CALCIUM ION, CHLORIDE ION, Olei00960, ... | | 著者 | Singer, A.U, Kagan, O, Kim, Y, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-02-25 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.998 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LMB
 
 | | The crystal structure of the protein OLEI01261 with unknown function from Chlorobaculum tepidum TLS | | 分子名称: | Uncharacterized protein | | 著者 | Zhang, R, Evdokimova, E, Egorova, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-29 | | 公開日 | 2010-03-16 | | 最終更新日 | 2013-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3M16
 
 | | Structure of a Transaldolase from Oleispira antarctica | | 分子名称: | Transaldolase | | 著者 | Singer, A.U, Kagan, O, Zhang, R, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-04 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3IRU
 
 | | Crystal structure of phoshonoacetaldehyde hydrolase like protein from Oleispira antarctica | | 分子名称: | SODIUM ION, phoshonoacetaldehyde hydrolase like protein | | 著者 | Chang, C, Evdokimova, E, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-08-24 | | 公開日 | 2009-09-01 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3LNP
 
 | | Crystal Structure of Amidohydrolase family Protein OLEI01672_1_465 from Oleispira antarctica | | 分子名称: | ACETIC ACID, Amidohydrolase family Protein OLEI01672_1_465, CALCIUM ION, ... | | 著者 | Kim, Y, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-02-02 | | 公開日 | 2010-02-16 | | 最終更新日 | 2013-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3S8Y
 
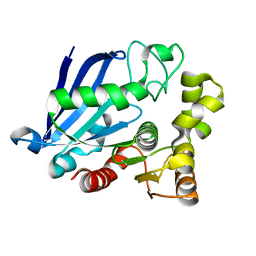 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | 分子名称: | BROMIDE ION, Esterase APC40077 | | 著者 | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | 登録日 | 2011-05-31 | | 公開日 | 2011-06-15 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3I4Q
 
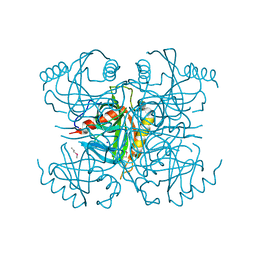 | | Structure of a putative inorganic pyrophosphatase from the oil-degrading bacterium Oleispira antarctica | | 分子名称: | APC40078, SODIUM ION | | 著者 | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2009-07-02 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Genome sequence and functional genomic analysis of the oil-degrading bacterium Oleispira antarctica.
Nat Commun, 4, 2013
|
|
3D1R
 
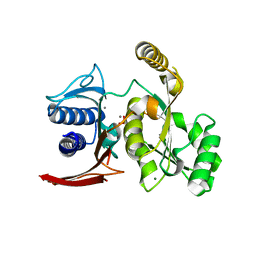 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-05-06 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3ERP
 
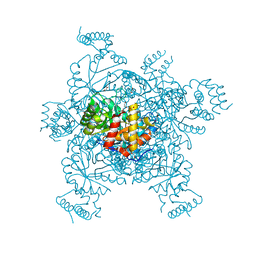 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | 分子名称: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-02 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
3EFV
 
 | | Crystal Structure of a Putative Succinate-Semialdehyde Dehydrogenase from Salmonella typhimurium LT2 with bound NAD | | 分子名称: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative succinate-semialdehyde dehydrogenase | | 著者 | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-09-10 | | 公開日 | 2008-11-04 | | 最終更新日 | 2013-06-12 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
3ETF
 
 | | Crystal structure of a putative succinate-semialdehyde dehydrogenase from salmonella typhimurium lt2 | | 分子名称: | Putative succinate-semialdehyde dehydrogenase | | 著者 | Brunzelle, J.S, Evdokimova, E, Kudritska, M, Wawrzak, Z, Anderson, W.F, Savchenk, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2008-10-07 | | 公開日 | 2008-11-04 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure and activity of the NAD(P)(+) -dependent succinate semialdehyde dehydrogenase YneI from Salmonella typhimurium.
Proteins, 81, 2013
|
|
6AQE
 
 | | Crystal structure of PPK2 in complex with Mg ATP | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | 著者 | Nocek, B, Joachimiak, A, Yakunin, A. | | 登録日 | 2017-08-19 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.805 Å) | | 主引用文献 | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
6AQN
 
 | | Crystal structure of PPK2 in complex with phosphonic acid inhibitor | | 分子名称: | GLYCEROL, Polyphosphate:AMP phosphotransferase, S,R MESO-TARTARIC ACID, ... | | 著者 | Nocek, B, Berlicki, l, Joachimiak, A, Yakunin, S. | | 登録日 | 2017-08-20 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.199 Å) | | 主引用文献 | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
6B18
 
 | | Crystal structure of PPK3 Class III in complex with inhibitor | | 分子名称: | GLYCEROL, PHOSPHATE ION, PPK3 Class III, ... | | 著者 | Nocek, B, Ruszkowski, M, Berlicki, L, Joachimiak, A, Yakunin, A. | | 登録日 | 2017-09-17 | | 公開日 | 2019-01-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural Insights into Substrate Selectivity and Activity of Bacterial Polyphosphate Kinases
Acs Catalysis, 8, 2018
|
|
6ANH
 
 | |
