3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | 分子名称: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | 著者 | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | 登録日 | 2010-12-13 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
5D6S
 
 | | Structure of epoxyqueuosine reductase from Streptococcus thermophilus. | | 分子名称: | COBALAMIN, Epoxyqueuosine reductase, IRON/SULFUR CLUSTER | | 著者 | Payne, K.A.P, Fisher, K, Dunstan, M.S, Sjuts, H, Leys, D. | | 登録日 | 2015-08-12 | | 公開日 | 2015-09-23 | | 最終更新日 | 2015-11-25 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Epoxyqueuosine Reductase Structure Suggests a Mechanism for Cobalamin-dependent tRNA Modification.
J.Biol.Chem., 290, 2015
|
|
5E1W
 
 | |
5E1Z
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,4-dichlorophenol bound form | | 分子名称: | 2,4-dichlorophenol, Transcriptional regulator, MarR family | | 著者 | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2015-09-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
to be published
|
|
5E20
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 2,3-dichlorophenol bound form | | 分子名称: | 2,3-dichlorophenol, Transcriptional regulator, MarR family | | 著者 | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2015-09-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
To be published
|
|
5E1X
 
 | | Crystal structure of the organohalide sensing RdhR-CbdbA1625 transcriptional regulator in the 3,4-dichlorophenol bound form | | 分子名称: | 3,4-dichlorophenol, Transcriptional regulator, MarR family | | 著者 | Quezada, C.P, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2015-09-30 | | 公開日 | 2016-10-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal structures of RdhRCbdbA1625 provide insight into sensing of chloroaromatic compounds by MarR-type regulators
TO BE PUBLISHED
|
|
4O1E
 
 | | Structure of a methyltransferase component in complex with MTHF involved in O-demethylation | | 分子名称: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, Dihydropteroate synthase DHPS | | 著者 | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2013-12-15 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O0Q
 
 | | Apo structure of a methyltransferase component involved in O-demethylation | | 分子名称: | Dihydropteroate synthase DHPS | | 著者 | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2013-12-14 | | 公開日 | 2015-01-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4O1F
 
 | | Structure of a methyltransferase component in complex with THF involved in O-demethylation | | 分子名称: | (6S)-5,6,7,8-TETRAHYDROFOLATE, Dihydropteroate synthase DHPS | | 著者 | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2013-12-15 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of the methyltransferase component of Desulfitobacterium hafniense DCB-2 O-demethylase shed light on methyltetrahydrofolate formation
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4JGI
 
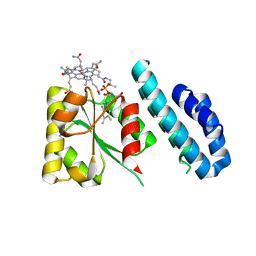 | | 1.5 Angstrom crystal structure of a novel cobalamin-binding protein from Desulfitobacterium hafniense DCB-2 | | 分子名称: | CO-METHYLCOBALAMIN, Putative uncharacterized protein | | 著者 | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | 登録日 | 2013-03-01 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5W4W
 
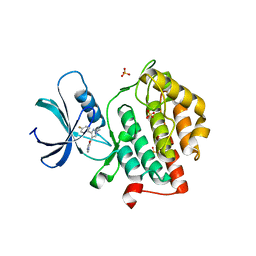 | |
5NY5
 
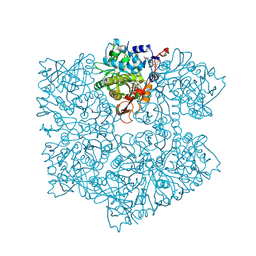 | | The apo structure of 3,4-dihydroxybenzoic acid decarboxylases from Enterobacter cloacae | | 分子名称: | 3,4-dihydroxybenzoate decarboxylase, GLYCEROL | | 著者 | Dordic, A, Gruber, K, Payer, S, Glueck, S, Pavkov-Keller, T, Marshall, S, Leys, D. | | 登録日 | 2017-05-11 | | 公開日 | 2017-09-13 | | 最終更新日 | 2020-11-18 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4KBC
 
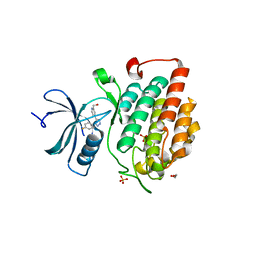 | |
4KB8
 
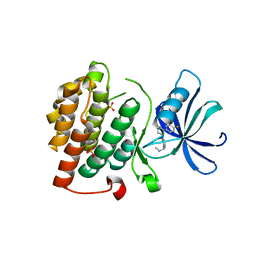 | |
5O3N
 
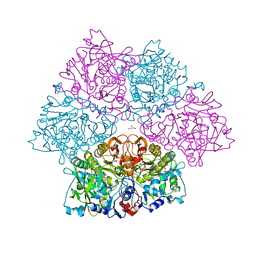 | | Crystal structure of E. cloacae 3,4-dihydroxybenzoic acid decarboxylase (AroY) reconstituted with prFMN | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-yl)-D-ribitol, 3,4-dihydroxybenzoate decarboxylase, GLYCEROL, ... | | 著者 | Marshall, S.A, Leys, D. | | 登録日 | 2017-05-24 | | 公開日 | 2017-09-13 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Regioselective para-Carboxylation of Catechols with a Prenylated Flavin Dependent Decarboxylase.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5O3M
 
 | |
4KBK
 
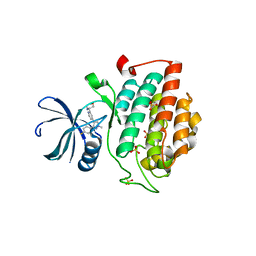 | |
4KBA
 
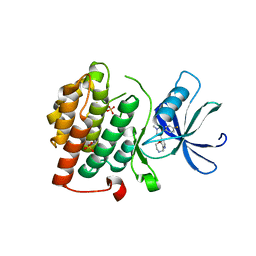 | | CK1d in complex with 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine inhibitor | | 分子名称: | 9-[3-(4-fluorophenyl)-1-methyl-1H-pyrazol-4-yl]-2,3,4,5-tetrahydropyrido[2,3-f][1,4]oxazepine, Casein kinase I isoform delta, SULFATE ION | | 著者 | Liu, S. | | 登録日 | 2013-04-23 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Ligand-protein interactions of selective casein kinase 1 delta inhibitors.
J.Med.Chem., 56, 2013
|
|
7P9Q
 
 | | Crystal structure of Indole 3-Carboxylic acid decarboxylase from Arthrobacter nicotianae FI1612 in complex with co-factor prFMN. | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, AnInD, MANGANESE (II) ION, ... | | 著者 | Gahloth, D, Leys, D. | | 登録日 | 2021-07-27 | | 公開日 | 2022-03-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.53 Å) | | 主引用文献 | Structural and biochemical characterization of the prenylated flavin mononucleotide-dependent indole-3-carboxylic acid decarboxylase.
J.Biol.Chem., 298, 2022
|
|
4RAS
 
 | | Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation | | 分子名称: | CHLORIDE ION, COBALAMIN, IRON/SULFUR CLUSTER, ... | | 著者 | Quezada, C.P, Payne, K.A.P, Leys, D. | | 登録日 | 2014-09-11 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation.
Nature, 517, 2015
|
|
5LTE
 
 | |
5M1E
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | 分子名称: | (16~{R})-11,12,14,14-tetramethyl-3,5-bis(oxidanylidene)-8-[(2~{S},3~{S},4~{R})-2,3,4-tris(oxidanyl)-5-phosphonooxy-pentyl]-1,4,6,8-tetrazatetracyclo[7.7.1.0^{2,7}.0^{13,17}]heptadeca-2(7),9(17),10,12-tetraene-16-sulfonic acid, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | 著者 | Marshall, S.A, Leys, D. | | 登録日 | 2016-10-07 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5LTI
 
 | |
5M1D
 
 | | Crystal structure of N-terminally tagged UbiD from E. coli reconstituted with prFMN cofactor | | 分子名称: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, 3-octaprenyl-4-hydroxybenzoate carboxy-lyase, MANGANESE (II) ION, ... | | 著者 | Marshall, S.A, Leys, D. | | 登録日 | 2016-10-07 | | 公開日 | 2017-01-11 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Oxidative Maturation and Structural Characterization of Prenylated FMN Binding by UbiD, a Decarboxylase Involved in Bacterial Ubiquinone Biosynthesis.
J. Biol. Chem., 292, 2017
|
|
5LTH
 
 | |
