4CMH
 
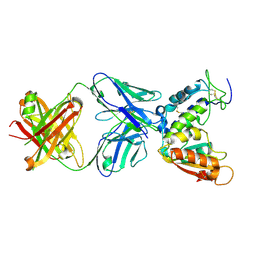 | | Crystal structure of CD38 with a novel CD38-targeting antibody SAR650984 | | Descriptor: | ADP-RIBOSYL CYCLASE 1, HEAVY CHAIN OF SAR650984-FAB FRAGMENT, LIGHT CHAIN OF SAR650984-FAB FRAGMENT | | Authors: | Deckert, J, Wetzel, M.C, Park, P.U, Bartle, L.M, Skaletskaya, A, Goldmacher, V, Vallee, F, ZhouLiu, Q, Ferrari, P, Pouzieux, S, Lahoute, C, Dumontet, C, Plesa, A, Chiron, M, Lejeune, P, Chittenden, T, Blanc, V. | | Deposit date: | 2014-01-15 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | SAR650984, a novel humanized CD38-targeting antibody, demonstrates potent antitumor activity in models of multiple myeloma and other CD38+ hematologic malignancies.
Clin. Cancer Res., 20, 2014
|
|
1Q42
 
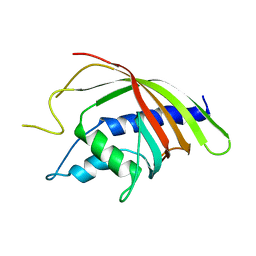 | | Crystal structure analysis of the Candida albicans Mtr2 | | Descriptor: | MRNA TRANSPORT REGULATOR Mtr2 | | Authors: | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | Deposit date: | 2003-08-01 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
1Q40
 
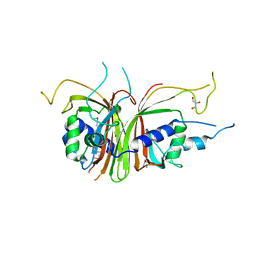 | | Crystal structure of the C. albicans Mtr2-Mex67 M domain complex | | Descriptor: | GLYCEROL, MRNA TRANSPORT REGULATOR Mtr2, mRNA export factor MEX67 | | Authors: | Senay, C, Ferrari, P, Rocher, C, Rieger, K.J, Winter, J, Platel, D, Bourne, Y. | | Deposit date: | 2003-08-01 | | Release date: | 2003-12-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The mtr2-mex67 ntf2-like domain complex: Structural
insights into a dual role of MTR2 for yeast nuclear export
J.Biol.Chem., 278, 2003
|
|
6O8D
 
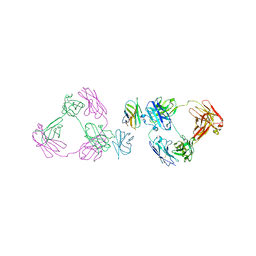 | | Anti-CD28xCD3 CODV Fab bound to CD28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-CD28xCD3 CODV Fab Heavy chain, ... | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2019-03-09 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Trispecific antibodies enhance the therapeutic efficacy of tumor-directed T cells through T cell receptor co-stimulation
To Be Published
|
|
8S9G
 
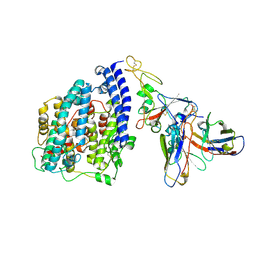 | | SARS-CoV-2 BN.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Fab Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-03-28 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
7U0A
 
 | |
7U0E
 
 | |
7U09
 
 | |
5FHX
 
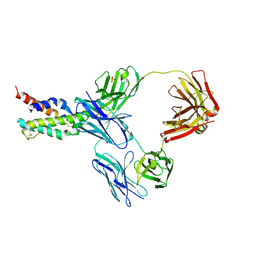 | | CRYSTAL STRUCTURE OF CODV IN COMPLEX WITH IL4 AT 2.55 Ang. RESOLUTION. | | Descriptor: | Antibody fragment light chain, Interleukin-4, PHOSPHATE ION, ... | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-30 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
5HCG
 
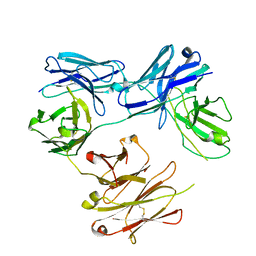 | | CRYSTAL STRUCTURE OF FREE-CODV. | | Descriptor: | CODV heavy-chain, CODV light chain | | Authors: | Vallee, F, Dupuy, A, Rak, A. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | CODV-Ig, a universal bispecific tetravalent and multifunctional immunoglobulin format for medical applications.
Mabs, 8, 2016
|
|
7SL5
 
 | |
7SKZ
 
 | |
8ERR
 
 | |
8ERQ
 
 | |
8FXB
 
 | | SARS-CoV-2 XBB.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
8FXC
 
 | | SARS-CoV-2 BQ.1.1 spike RBD bound to the human ACE2 ectodomain and the S309 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, S309 Heavy chain, ... | | Authors: | Park, Y.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2023-01-24 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Neutralization, effector function and immune imprinting of Omicron variants.
Nature, 621, 2023
|
|
6I9J
 
 | | Human transforming growth factor beta2 in a tetragonal crystal form | | Descriptor: | Transforming growth factor beta-2 proprotein | | Authors: | Gomis-Ruth, F.X, Marino-Puertas, L, del Amo-Maestro, L, Goulas, T. | | Deposit date: | 2018-11-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recombinant production, purification, crystallization, and structure analysis of human transforming growth factor beta 2 in a new conformation.
Sci Rep, 9, 2019
|
|
6O89
 
 | | Anti-CD28xCD3 CODV Fab | | Descriptor: | Anti-CD28xCD3 CODV-Fab Heavy chain, Anti-CD28xCD3 CODV-Fab Light chain | | Authors: | Lord, D.M, Wei, R.R. | | Deposit date: | 2019-03-09 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Trispecific antibodies enhance the therapeutic efficacy of tumor-directed T cells through T cell receptor co-stimulation
Nat Cancer, 1, 2020
|
|
7RNJ
 
 | | S2P6 Fab fragment bound to the SARS-CoV/SARS-CoV-2 spike stem helix peptide | | Descriptor: | Monoclonal antibody S2P6 Fab heavy chain, Monoclonal antibody S2P6 Fab light chain, SULFATE ION, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M, Sauer, M.M, Veesler, D. | | Deposit date: | 2021-07-29 | | Release date: | 2021-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Broad betacoronavirus neutralization by a stem helix-specific human antibody.
Science, 373, 2021
|
|
4KXZ
 
 | | crystal structure of tgfb2 in complex with GC2008. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, GC1008 Heavy Chain, ... | | Authors: | Mathieu, M, Moulin, A.G, Wei, R. | | Deposit date: | 2013-05-28 | | Release date: | 2014-09-24 | | Last modified: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
4KV5
 
 | | scFv GC1009 in complex with TGF-beta1. | | Descriptor: | Single-chain variable fragment GC1009, Transforming growth factor beta-1 proprotein | | Authors: | Wei, R, Moulin, A.G, Mathieu, M. | | Deposit date: | 2013-05-22 | | Release date: | 2014-09-24 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of a pan-specific antagonist antibody complexed to different isoforms of TGF beta reveal structural plasticity of antibody-antigen interactions.
Protein Sci., 23, 2014
|
|
2YJW
 
 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 4-(5-METHYL-4-PHENYLISOXAZOL-3-YL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-24 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YKJ
 
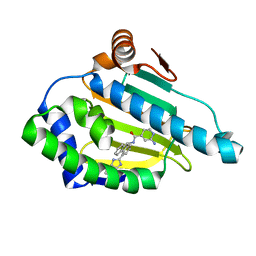 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 2-AMINO-N-[4-(3H-IMIDAZO[4,5-C]PYRIDIN-2-YL)--9H-FLUOREN-9-YL]-ISONICOTINAMIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
2YK2
 
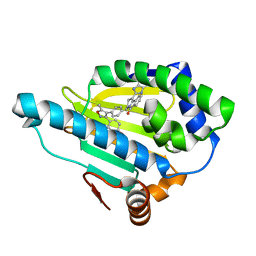 | | Tricyclic series of Hsp90 inhibitors | | Descriptor: | 1-(3H-imidazo[4,5-c]pyridin-2-yl)-3,4-dihydropyrido[2,1-a]isoindol-6(2H)-one, 4-(5-METHYL-4-PHENYLISOXAZOL-3-YL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Dupuy, A, Vallee, F. | | Deposit date: | 2011-05-25 | | Release date: | 2011-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Tricyclic Series of Heat Shock Protein 90 (Hsp90) Inhibitors Part I: Discovery of Tricyclic Imidazo[4,5-C]Pyridines as Potent Inhibitors of the Hsp90 Molecular Chaperone.
J.Med.Chem., 54, 2011
|
|
4UAG
 
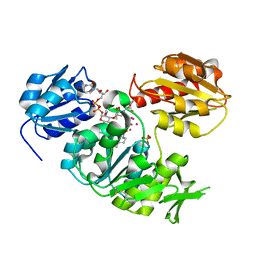 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, UNKNOWN ATOM OR ION, ... | | Authors: | Bertrand, J, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-03-09 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
