3VE6
 
 | |
1AYJ
 
 | |
1BK8
 
 | |
2JBV
 
 | | Crystal structure of choline oxidase reveals insights into the catalytic mechanism | | Descriptor: | CHOLINE OXIDASE, DIMETHYL SULFOXIDE, UNKNOWN ATOM OR ION, ... | | Authors: | Lountos, G.T, Fan, F, Gadda, G, Orville, A.M. | | Deposit date: | 2006-12-13 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Role of Glu312 in Binding and Positioning of the Substrate for the Hydride Transfer Reaction in Choline Oxidase.
Biochemistry, 47, 2008
|
|
3HGL
 
 | | crystal of AvrPtoB 121-205 | | Descriptor: | Effector protein hopAB2 | | Authors: | Dong, J, Xiao, F, Fan, F, Gu, L, Cang, H, Martin, G.B, Chai, J. | | Deposit date: | 2009-05-14 | | Release date: | 2009-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Complex between Pseudomonas Effector AvrPtoB and the Tomato Pto Kinase Reveals Both a Shared and a Unique Interface Compared with AvrPto-Pto
Plant Cell, 21, 2009
|
|
3HGK
 
 | | crystal structure of effect protein AvrptoB complexed with kinase Pto | | Descriptor: | Effector protein hopAB2, Protein kinase | | Authors: | Dong, J, Fan, F, Gu, L, Chai, J. | | Deposit date: | 2009-05-14 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the Complex between Pseudomonas Effector AvrPtoB and the Tomato Pto Kinase Reveals Both a Shared and a Unique Interface Compared with AvrPto-Pto
Plant Cell, 21, 2009
|
|
3C8Z
 
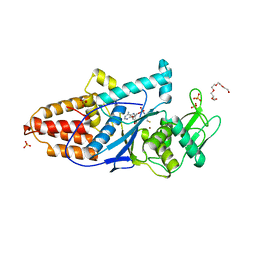 | | The 1.6 A Crystal Structure of MshC: The Rate Limiting Enzyme in the Mycothiol Biosynthetic Pathway | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, Cysteinyl-tRNA synthetase, ... | | Authors: | Tremblay, L.W, Fan, F, Vetting, M.W, Blanchard, J.S. | | Deposit date: | 2008-02-14 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of Mycobacterium smegmatis MshC: the penultimate enzyme in the mycothiol biosynthetic pathway.
Biochemistry, 47, 2008
|
|
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
5XMK
 
 | | Cryo-EM structure of the ATP-bound Vps4 mutant-E233Q complex with Vta1 (masked) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | Deposit date: | 2017-05-15 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
3M6B
 
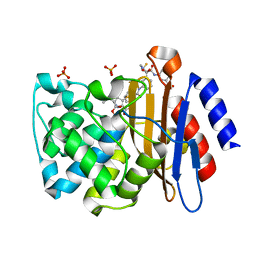 | | Crystal Structure of the Ertapenem Pre-isomerized Covalent Adduct with TB B-lactamase | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Fan, F, Blanchard, J.S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
6IP1
 
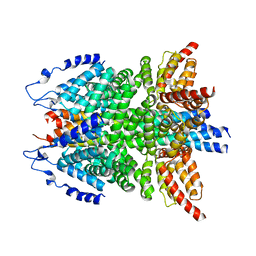 | | alpha-SNAP-SNARE subcomplex in the whole 20S complex | | Descriptor: | Alpha-soluble NSF attachment protein, Synaptosomal-associated protein 25, Syntaxin-1A, ... | | Authors: | Huang, X, Sun, S, Wang, X, Fan, F, Zhou, Q, Sui, S.F. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanistic insights into the SNARE complex disassembly.
Sci Adv, 5, 2019
|
|
6IP2
 
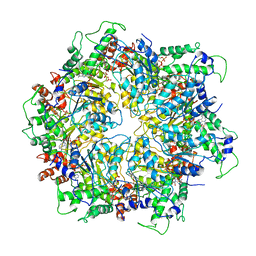 | | NSF-D1D2 part in the whole 20S complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Vesicle-fusing ATPase | | Authors: | Huang, X, Sun, S, Wang, X, Fan, F, Zhou, Q, Sui, S.F. | | Deposit date: | 2018-11-01 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanistic insights into the SNARE complex disassembly.
Sci Adv, 5, 2019
|
|
1WB7
 
 | | Iron Superoxide Dismutase (Fe-SOD) From The Hyperthermophile Sulfolobus Solfataricus. Crystal Structure of the Y41F mutant. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Gogliettino, M.A, Tanfani, F, Scire, A, Ursby, T, Adinolfi, B.S, Cacciamani, T, De Vendittis, E. | | Deposit date: | 2004-10-31 | | Release date: | 2004-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Role of Tyr41 and His155 in the Functional Properties of Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus
Biochemistry, 43, 2004
|
|
3HXP
 
 | | Crystal structure of the FhuD fold-family BSU3320, a periplasmic binding protein component of a Fep/Fec-like ferrichrome ABC transporter from Bacillus subtilis. Northeast Structural Genomics Consortium Target SR577 | | Descriptor: | Iron(3+)-hydroxamate-binding protein fhuD | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-21 | | Release date: | 2009-07-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR577
To be Published
|
|
3LM6
 
 | | Crystal Structure of Stage V sporulation protein AD (spoVAD) from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR525 | | Descriptor: | Stage V sporulation protein AD | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR525
To be Published
|
|
3GUW
 
 | | Crystal Structure of the TatD-like Protein (AF1765) from Archaeoglobus fulgidus, Northeast Structural Genomics Consortium Target GR121 | | Descriptor: | ZINC ION, uncharacterized protein AF_1765 | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-30 | | Release date: | 2009-04-07 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target GR121
To be Published
|
|
3GVZ
 
 | | Crystal structure of the protein CV2077 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR62 | | Descriptor: | Uncharacterized protein CV2077 | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Maglaqui, M, Owens, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-14 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Protein CV2077 from Chromobacterium violaceum. Northeast Structural Genomics Consortium Target CvR62.
To be Published
|
|
3GOC
 
 | | Crystal structure of the Endonuclease V (SAV1684) from Streptomyces avermitilis. Northeast Structural Genomics Consortium Target SvR196 | | Descriptor: | 3-(2-hydroxyethyl)-2,2-bis(hydroxymethyl)pentane-1,5-diol, CHLORIDE ION, Endonuclease V, ... | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Owens, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the Endonuclease V (SAV1684) from Streptomyces avermitilis. Northeast Structural Genomics Consortium Target SvR196.
To be Published
|
|
1UNC
 
 | | Solution structure of the human villin C-terminal headpiece subdomain | | Descriptor: | VILLIN 1 | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
1UND
 
 | | Solution structure of the human advillin C-terminal headpiece subdomain | | Descriptor: | ADVILLIN | | Authors: | Vermeulen, W, Van Troys, M, Vanhaesebrouck, P, Verschueren, M, Fant, F, Ampe, C, Martins, J, Borremans, F. | | Deposit date: | 2003-09-09 | | Release date: | 2004-07-15 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of the C-Terminal Headpiece Subdomains of Human Villin and Advillin, Evaluation of Headpiece F-Actin-Binding Requirements
Protein Sci., 13, 2004
|
|
1CE4
 
 | |
3KVP
 
 | | Crystal Structure of Uncharacterized protein ymzC Precursor from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR378A | | Descriptor: | ACETIC ACID, Uncharacterized protein ymzC | | Authors: | Kuzin, A.P, Chen, Y, Seetharaman, J, Afonine, P, Fang, F, Xiao, R, Cunningham, K, Ma, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-30 | | Release date: | 2010-02-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR378A
To be Published
|
|
3GWB
 
 | | Crystal structure of peptidase M16 inactive domain from Pseudomonas fluorescens. Northeast Structural Genomics target PlR293L | | Descriptor: | Peptidase M16 inactive domain family protein | | Authors: | Seetharaman, J, Chen, Y, Fang, F, Xiao, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-31 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of peptidase M16 inactive domain from Pseudomonas fluorescens. Northeast Structural Genomics target PlR293L
To be Published
|
|
4H9D
 
 | | Crystal Structure of Mn-dependent Gme HNH nicking endonuclease from Geobacter metallireducens GS-15, Northeast Structural Genomics Consortium (NESG) Target GmR87 | | Descriptor: | HNH endonuclease, MAGNESIUM ION, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Owens, L, Chen, C.X, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-10 | | Method: | X-RAY DIFFRACTION (2.599 Å) | | Cite: | Northeast Structural Genomics Consortium Target GmR87
To be Published
|
|
2ZMV
 
 | | Crystal structure of Synbindin | | Descriptor: | Trafficking protein particle complex subunit 4 | | Authors: | Fan, F. | | Deposit date: | 2008-04-21 | | Release date: | 2008-07-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human synbindin reveals two conformations of longin domain
Biochem.Biophys.Res.Commun., 378, 2009
|
|
