5MW2
 
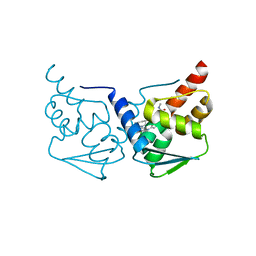 | | CRYSTAL STRUCTURE OF BCL-6 BTB-domain with BI-3802 | | Descriptor: | 2-[6-[[5-chloranyl-2-[(3~{S},5~{R})-3,5-dimethylpiperidin-1-yl]pyrimidin-4-yl]amino]-1-methyl-2-oxidanylidene-quinolin-3-yl]oxy-~{N}-methyl-ethanamide, B-cell lymphoma 6 protein | | Authors: | Bader, G, Flotzinger, G, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2017-01-18 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chemically Induced Degradation of the Oncogenic Transcription Factor BCL6.
Cell Rep, 20, 2017
|
|
6MQN
 
 | |
6MQG
 
 | |
4ER5
 
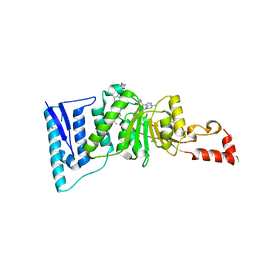 | | Crystal structure of human DOT1L in complex with 2 molecules of EPZ004777 | | Descriptor: | 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER3
 
 | | Crystal Structure of Human DOT1L in complex with inhibitor EPZ004777 | | Descriptor: | 1,2-ETHANEDIOL, 7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER6
 
 | | Crystal structure of human DOT1L in complex with inhibitor SGC0946 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, BROMIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
5FFO
 
 | | Integrin alpha V beta 6 in complex with pro-TGF-beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Zhao, B, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
5FFG
 
 | | Crystal structure of integrin alpha V beta 6 head | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2015-12-18 | | Release date: | 2017-01-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Force interacts with macromolecular structure in activation of TGF-beta.
Nature, 542, 2017
|
|
3IKA
 
 | |
3K5V
 
 | | Structure of Abl kinase in complex with imatinib and GNF-2 | | Descriptor: | 3-(6-{[4-(trifluoromethoxy)phenyl]amino}pyrimidin-4-yl)benzamide, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, ... | | Authors: | Cowan-Jacob, S.W, Fendrich, G, Rummel, G, Strauss, A. | | Deposit date: | 2009-10-08 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors.
Nature, 463, 2010
|
|
4EQZ
 
 | | Crystal structure of human DOT1L in complex with inhibitor FED2 | | Descriptor: | 5'-deoxy-5'-[(3-{[(4-methylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]adenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER0
 
 | | Crystal Structure of human DOT1L in complex with inhibitor FED1 | | Descriptor: | 5'-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)(propan-2-yl)amino]-5'-deoxyadenosine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Li, Y, Nguyen, K.T, Federation, A, Marineau, J, Qi, J, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
4ER7
 
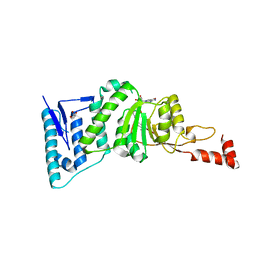 | | Crystal Structure of human DOT1L in complex with inhibitor SGC0947 | | Descriptor: | 5-bromo-7-{5-[(3-{[(4-tert-butylphenyl)carbamoyl]amino}propyl)amino]-5-deoxy-beta-D-ribofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, GLYCEROL, Histone-lysine N-methyltransferase, ... | | Authors: | Wernimont, A.K, Tempel, W, Yu, W, Scopton, A, Li, Y, Nguyen, K.T, Vedadi, M, Bradner, J.E, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-19 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors.
Nat Commun, 3, 2012
|
|
7QO6
 
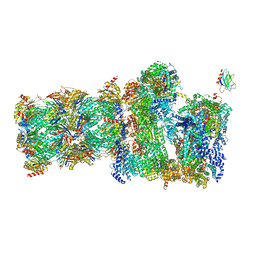 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO5
 
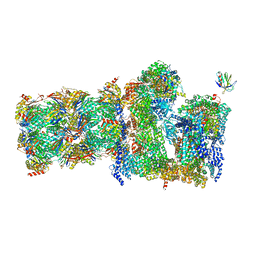 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO3
 
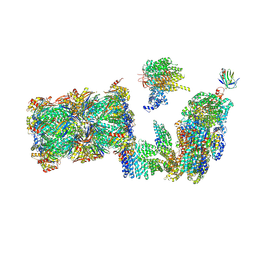 | | Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid) | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO4
 
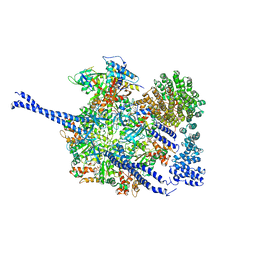 | | 26S proteasome WT-Ubp6-UbVS complex in the si state (ATPases, Rpn1, Ubp6, and UbVS) | | Descriptor: | 26S proteasome regulatory subunit RPN1, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
2N3T
 
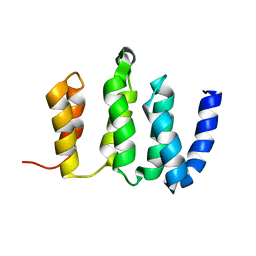 | |
2N3W
 
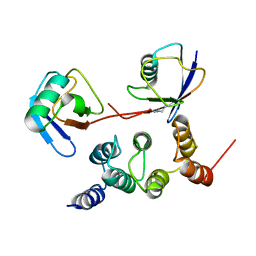 | |
2N3U
 
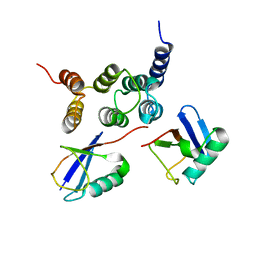 | |
2N3V
 
 | |
6OM1
 
 | | Crystal structure of an atypical integrin | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.C, Springer, T.A. | | Deposit date: | 2019-04-17 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | General structural features that regulate integrin affinity revealed by atypical alpha V beta 8.
Nat Commun, 10, 2019
|
|
6OM2
 
 | |
6V4E
 
 | | Crystal Structure Analysis of Zebra Fish MDM | | Descriptor: | Protein Mdm4, Stapled peptide QSQQTF(0EH)NLWRLL(MK8)QN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
6V4G
 
 | | Crystal Structure Analysis of Zebra fish MDMX | | Descriptor: | Protein Mdm4, SULFATE ION, Stapled peptide QSQQTF(0EH)NLWRLE(MK8)QN(NH2) | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-11-27 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Identification of a Structural Determinant for Selective Targeting of HDMX.
Structure, 28, 2020
|
|
