1G7O
 
 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
1SSU
 
 | | Structural and biochemical evidence for disulfide bond heterogeneity in active forms of the somatomedin B domain of human vitronectin | | Descriptor: | Vitronectin | | Authors: | Kamikubo, Y, De Guzman, R, Kroon, G, Curriden, S, Neels, J.G, Churchill, M.J, Dawson, P, Oldziej, S, Jagielska, A, Scheraga, H.A, Loskutoff, D.J, Dyson, H.J. | | Deposit date: | 2004-03-24 | | Release date: | 2004-07-27 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Disulfide bonding arrangements in active forms of the somatomedin B domain of human vitronectin.
Biochemistry, 43, 2004
|
|
1U2N
 
 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
1TOT
 
 | | ZZ Domain of CBP- a Novel Fold for a Protein Interaction Module | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Legge, G.B, Martinez-Yamout, M.A, Hambly, D.M, Trinh, T, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-06-15 | | Release date: | 2005-01-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module
J.Mol.Biol., 343, 2004
|
|
1XJH
 
 | | NMR structure of the redox switch domain of the E. coli Hsp33 | | Descriptor: | 33 kDa chaperonin, ZINC ION | | Authors: | Won, H.S, Low, L.Y, De Guzman, R.N, Martinez-Yamout, M.A, Jakob, U, Dyson, H.J. | | Deposit date: | 2004-09-23 | | Release date: | 2004-10-05 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The Zinc-dependent Redox Switch Domain of the Chaperone Hsp33 has a Novel Fold
J.Mol.Biol., 341, 2004
|
|
2M9F
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9E
 
 | | NMR solution structure of Pin1 WW domain mutant 5-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-07 | | Release date: | 2013-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9J
 
 | | NMR solution structure of Pin1 WW domain mutant 6-1g | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2M9I
 
 | | NMR solution structure of Pin1 WW domain variant 6-1 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Enck, S, Chen, W, Price, J.L, Powers, E.T, Wong, C, Dyson, H.J, Kelly, J.W. | | Deposit date: | 2013-06-10 | | Release date: | 2013-06-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins.
J.Am.Chem.Soc., 135, 2013
|
|
2PRT
 
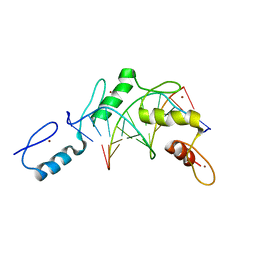 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*AP*CP*GP*CP*CP*CP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*GP*GP*GP*CP*GP*TP*CP*TP*G)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the Wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2HDP
 
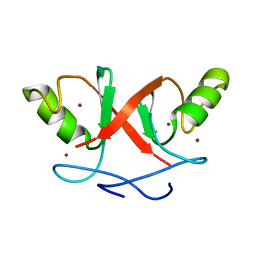 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
2JP9
 
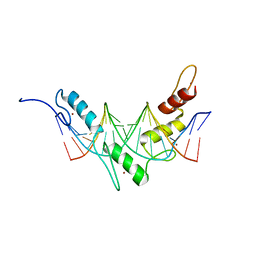 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JPA
 
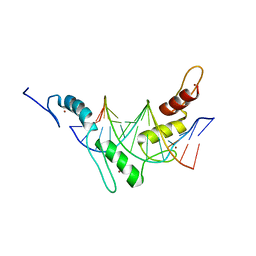 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2020-02-05 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2KA4
 
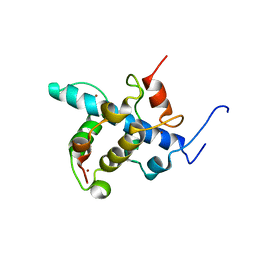 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
2KA6
 
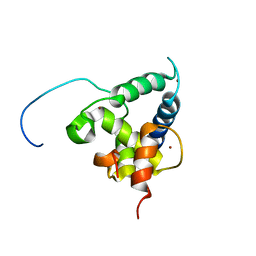 | | NMR structure of the CBP-TAZ2/STAT1-TAD complex | | Descriptor: | CREB-binding protein, Signal transducer and activator of transcription 1-alpha/beta, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-07 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains.
Embo J., 28, 2009
|
|
2L14
 
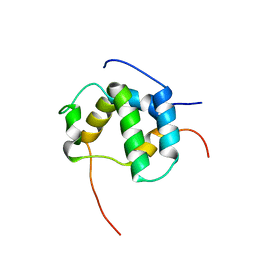 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
8W2W
 
 | |
8W1N
 
 | |
1AI1
 
 | | HIV-1 V3 LOOP MIMIC | | Descriptor: | AIB142, IGG1-KAPPA 59.1 FAB (HEAVY CHAIN), IGG1-KAPPA 59.1 FAB (LIGHT CHAIN) | | Authors: | Ghiara, J.B, Wilson, I.A. | | Deposit date: | 1996-11-06 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site.
J.Mol.Biol., 266, 1997
|
|
8EDF
 
 | |
8ECZ
 
 | | Bovine Fab 4C1 | | Descriptor: | 4C1 Fab heavy chain, 4C1 Fab light chain, PHOSPHATE ION | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECQ
 
 | | Bovine Fab 2G3 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2G3 Fab Heavy chain, 2G3 Fab Light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ECV
 
 | | Bovine Fab 2F12 | | Descriptor: | 2F12 Fab Heavy chain, 2F12 Fab Light chain | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ED1
 
 | | Bovine Fab 5C1 | | Descriptor: | 5C1 Fab heavy chain, 5C1 Fab light chain, GLYCEROL, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-09-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The smallest functional antibody fragment: Ultralong CDR H3 antibody knob regions potently neutralize SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5CC9
 
 | |
