5IW5
 
 | |
5IW4
 
 | |
7W84
 
 | |
7W88
 
 | |
7W82
 
 | | Crystal structure of maize RDR2 | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Du, X, Yang, Z, Du, J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production.
Plant Cell, 34, 2022
|
|
5ZWZ
 
 | |
5ZWX
 
 | |
6NF0
 
 | | Nocturnin with bound NADPH substrate | | Descriptor: | CALCIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Nocturnin | | Authors: | Estrella, M.A, Du, J, Korennykh, A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The metabolites NADP+and NADPH are the targets of the circadian protein Nocturnin (Curled).
Nat Commun, 10, 2019
|
|
7SQ1
 
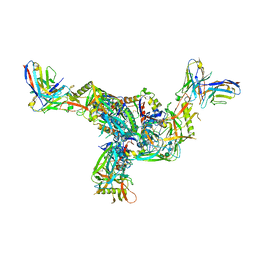 | | BG505.MD39TS Env trimer in complex with Fab from antibody C05 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C05 Fab Light chain, ... | | Authors: | Moore, A, Du, J, Xu, Z, Walker, S, Kulp, D.W, Pallesen, J. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-22 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Induction of tier-2 neutralizing antibodies in mice with a DNA-encoded HIV envelope native like trimer.
Nat Commun, 13, 2022
|
|
8HIL
 
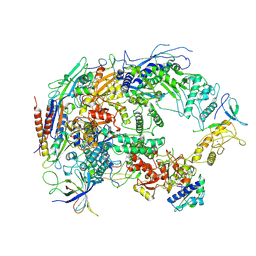 | | A cryo-EM structure of B. oleracea RNA polymerase V at 3.57 Angstrom | | Descriptor: | DNA-dependent RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, DNA-directed RNA polymerase subunit, ... | | Authors: | Du, X, Xie, G, Hu, H, Du, J. | | Deposit date: | 2022-11-20 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
8HIM
 
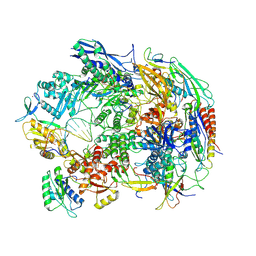 | | A cryo-EM structure of B. oleracea RNA polymerase V elongation complex at 2.73 Angstrom | | Descriptor: | DNA (34-MER), DNA-directed RNA polymerase IV and V subunit 2, DNA-directed RNA polymerase V largest subunit, ... | | Authors: | Hu, H, Xie, G, Du, X, Du, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and mechanism of the plant RNA polymerase V.
Science, 379, 2023
|
|
5IX2
 
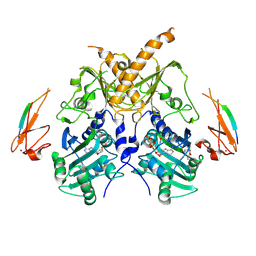 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and unmodified H3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4H0N
 
 | | Crystal structure of Spodoptera frugiperda DNMT2 E260A/E261A/K263A mutant | | Descriptor: | CALCIUM ION, DNMT2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Li, S, Du, J, Yang, H, Yin, J, Zhong, J, Ding, J. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Functional and structural characterization of DNMT2 from Spodoptera frugiperda.
J Mol Cell Biol, 5, 2013
|
|
5IX1
 
 | | Crystal structure of mouse Morc3 ATPase-CW cassette in complex with AMPPNP and H3K4me3 peptide | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 3, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, S, Du, J, Patel, D.J. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-17 | | Last modified: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3FGG
 
 | | Crystal Structure of Putative ECF-type Sigma Factor Negative Effector from Bacillus cereus | | Descriptor: | GLYCEROL, ZINC ION, uncharacterized protein BCE2196 | | Authors: | Kim, Y, Nocek, B, Maltseva, N, Joachimiak, G, Du, J, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Putative ECF-type Sigma Factor Negative Effector from Bacillus cereus
To be Published
|
|
3FH3
 
 | | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne | | Descriptor: | NICKEL (II) ION, putative ECF-type sigma factor negative effector | | Authors: | Nocek, B, Kim, Y, Joachimiak, G, Du, J, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2009-01-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Crystal structure of a putative ECF-type sigma factor negative effector from Bacillus anthracis str. Sterne
To be Published
|
|
7O12
 
 | | ABC transporter NosDFY, AMPPNP-bound in GDN | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O10
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O11
 
 | | ABC transporter NosDFY, nucleotide-free in GDN, R-domain 1 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O0Y
 
 | | ABC transporter NosDFY, nucleotide-free in GDN | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O0Z
 
 | | ABC transporter NosFY, nucleotide-free in GDN | | Descriptor: | Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter permease protein NosY | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O13
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O15
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 2 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O17
 
 | | ABC transporter NosDFY E154Q, ATP-bound in lipid nanodisc | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
7O16
 
 | | ABC transporter NosDFY, nucleotide-free in lipid nanodisc, R-domain 3 | | Descriptor: | MAGNESIUM ION, Probable ABC transporter ATP-binding protein NosF, Probable ABC transporter binding protein NosD, ... | | Authors: | Mueller, C, Zhang, L, Lu, W, Einsle, O, Du, J. | | Deposit date: | 2021-03-28 | | Release date: | 2022-04-13 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
