4FNV
 
 | | Crystal Structure of Heparinase III | | Descriptor: | Heparinase III protein, heparitin sulfate lyase | | Authors: | Dong, W, Ye, S. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of heparan sulfate-specific degradation by heparinase III.
Protein Cell, 3, 2012
|
|
5B6L
 
 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-05-30 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
4RO0
 
 | | Crystal structure of MthK gating ring in a ligand-free form | | Descriptor: | Calcium-gated potassium channel MthK | | Authors: | Dong, W, Guo, R, Chai, H, Chen, Z, Cui, H, Ren, Z, Li, Y, Ye, S. | | Deposit date: | 2014-10-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The principle of cooperative gating in a calcium-gated K+ channel
To be Published
|
|
5GND
 
 | | Structure of Deg protease HhoA from Synechocystis sp. PCC 6803 | | Descriptor: | Putative serine protease HhoA, SODIUM ION, UNK-UNK-UNK-UNK-TRP, ... | | Authors: | Dong, W, Wang, J, Liu, L. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the zinc-bound HhoA protease from Synechocystis sp. PCC 6803
Febs Lett., 590, 2016
|
|
4E44
 
 | |
4E45
 
 | | Crystal structure of the hMHF1/hMHF2 Histone-Fold Tetramer in Complex with Fanconi Anemia Associated Helicase hFANCM | | Descriptor: | Centromere protein S, Centromere protein X, Fanconi anemia group M protein, ... | | Authors: | Fox III, D, Zhao, Y, Yang, W, Weidong, W. | | Deposit date: | 2012-03-12 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal that FANCM remodels the MHF Tetramer in favor of binding Branched DNA
To be Published
|
|
4WZF
 
 | | Crystal structural basis for Rv0315, an immunostimulatory antigen and pseudo beta-1, 3-glucanase of Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanase, CALCIUM ION | | Authors: | Dong, W.Y, Fu, Z.F, Peng, G.Q. | | Deposit date: | 2014-11-19 | | Release date: | 2015-12-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structural basis for Rv0315, an immunostimulatory antigen and inactive beta-1,3-glucanase of Mycobacterium tuberculosis.
Sci Rep, 5, 2015
|
|
8R8Q
 
 | | Lysosomal peptide transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycosylated lysosomal membrane protein, Major facilitator superfamily domain-containing protein 1 | | Authors: | Jungnickel, K.E.J, Loew, C. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | MFSD1 with its accessory subunit GLMP functions as a general dipeptide uniporter in lysosomes.
Nat.Cell Biol., 26, 2024
|
|
4N7R
 
 | | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its binding protein | | Descriptor: | Genomic DNA, chromosome 3, P1 clone: MXL8, ... | | Authors: | Zhao, A, Fang, Y, Lin, Y, Gong, W, Liu, L. | | Deposit date: | 2013-10-16 | | Release date: | 2014-05-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal structure of Arabidopsis glutamyl-tRNA reductase in complex with its stimulator protein
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6C9N
 
 | |
6C67
 
 | | Mycobacterium tuberculosis adenosine kinase bound to iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9V
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | (2R,3S,4R,5R)-2-(hydroxymethyl)-5-[6-(4-phenylpiperazin-1-yl)-9H-purin-9-yl]tetrahydrofuran-3,4-diol, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9S
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3R,4S,5R)-2-(6-([1,1'-biphenyl]-4-ylethynyl)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 6-[([1,1'-biphenyl]-4-yl)ethynyl]-9-beta-D-ribofuranosyl-9H-purine, Adenosine kinase, SODIUM ION, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
6C9P
 
 | |
6C9Q
 
 | |
6C9R
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
8J1Z
 
 | | The global structure of pre50S related to DbpA in state3 | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yu, T, Zeng, F. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PTC Remodeling in Pre50S Intermediates: Insights into the Role of DEAD-box RNA Helicase DbpA
To Be Published
|
|
1RRP
 
 | | STRUCTURE OF THE RAN-GPPNHP-RANBD1 COMPLEX | | Descriptor: | MAGNESIUM ION, NUCLEAR PORE COMPLEX PROTEIN NUP358, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vetter, I.R, Nowak, C, Nishimoto, T, Kuhlmann, J, Wittinghofer, A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.
Nature, 398, 1999
|
|
7F3B
 
 | | cocrystallization of Escherichia coli dihydrofolate reductase (DHFR) and its pyrrolo[3,2-f]quinazoline inhibitor. | | Descriptor: | 7-[(2-fluorophenyl)methyl]pyrrolo[3,2-f]quinazoline-1,3-diamine, Dihydrofolate reductase, GLYCEROL | | Authors: | Wang, H, You, X.F, Yang, X.Y, Li, Y, Hong, W. | | Deposit date: | 2021-06-16 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The discovery of 1, 3-diamino-7H-pyrrol[3, 2-f]quinazoline compounds as potent antimicrobial antifolates.
Eur.J.Med.Chem., 228, 2022
|
|
4QS9
 
 | |
4QS8
 
 | |
4QS7
 
 | |
7DXO
 
 | |
7CKK
 
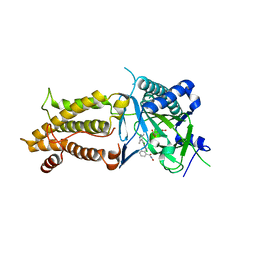 | | Structural complex of FTO bound with Dac51 | | Descriptor: | 2-{[2,6-dichloro-4-(3,5-dimethyl-1H-pyrazol-4-yl)phenyl]amino}-N-hydroxybenzamide, Alpha-ketoglutarate-dependent dioxygenase FTO, N-OXALYLGLYCINE | | Authors: | Yang, C, Gan, J. | | Deposit date: | 2020-07-17 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance.
Cell Metab., 33, 2021
|
|
7CRN
 
 | |
