8SA3
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA2
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
7M2N
 
 | | Crystal structure of Human Lactate Dehydrogenase A with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
7M2O
 
 | | Crystal structure of Human Glycolate Oxidase with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
1HPZ
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | ALPHA-(2,6-DICHLOROPHENYL)-ALPHA-(2-ACETYL-5-METHYLANILINO)ACETAMIDE, POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-13 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
1HQE
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | POL POLYPROTEIN | | Authors: | Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 2000-12-15 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
4YKC
 
 | |
4YKD
 
 | |
4YL6
 
 | |
3WA5
 
 | | Crystal Structure of type VI peptidoglycan muramidase effector Tse3 in complex with its cognate immunity protein Tsi3 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Tse3-specific immunity protein, ... | | Authors: | Ding, J, Wang, T, Liu, W, Wang, D.C. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Complex structure of type VI peptidoglycan muramidase effector and a cognate immunity protein.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WBM
 
 | | Crystal Structure of protein-RNA complex | | Descriptor: | DNA/RNA-binding protein Alba 1, RNA (25-MER) | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2013-05-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea.
J.Biol.Chem., 289, 2014
|
|
2HTG
 
 | | Structural and functional characterization of TM VII of the NHE1 isoform of the Na+/H+ exchanger | | Descriptor: | NHE1 isoform of Na+/H+ exchanger 1 | | Authors: | Rainey, J.K, Ding, J, Xu, C, Fliegel, L, Sykes, B.D. | | Deposit date: | 2006-07-25 | | Release date: | 2006-08-08 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structural and functional characterization of transmembrane segment VII of the Na(+)/H(+) exchanger isoform 1
J.Biol.Chem., 281, 2006
|
|
5Z2C
 
 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
7WZS
 
 | |
2MYJ
 
 | |
7DN8
 
 | | Crystal structure of Salmonella effector SopF in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2022-12-28 | | Method: | X-RAY DIFFRACTION (2.6084 Å) | | Cite: | ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7DN9
 
 | |
6KN1
 
 | | P20/P12 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-02 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KN0
 
 | |
6KMZ
 
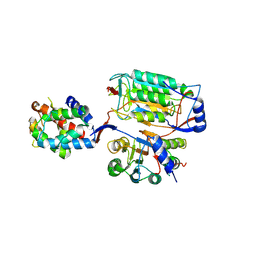 | |
6KMT
 
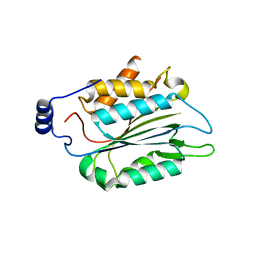 | | P32 of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMU
 
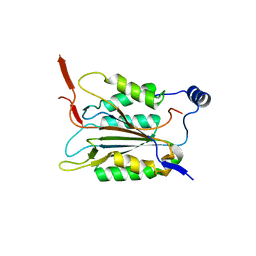 | | P22/P10 complex of caspase-11 mutant C254A | | Descriptor: | Caspase-4 | | Authors: | Ding, J, Sun, Q. | | Deposit date: | 2019-08-01 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Mechanism for GSDMD Targeting by Autoprocessed Caspases in Pyroptosis.
Cell, 180, 2020
|
|
6KMV
 
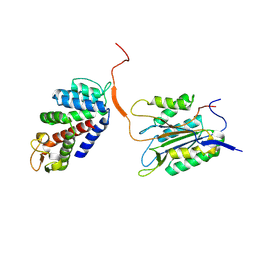 | |
