4D5E
 
 | | Crystal Structure of recombinant wildtype CDH | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Loschonsky, S, Wacker, T, Waltzer, S, Giovannini, P.P, McLeish, M.J, Andrade, S.L.A, Mueller, M. | | Deposit date: | 2014-11-03 | | Release date: | 2015-01-21 | | Last modified: | 2017-08-23 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Extended Reaction Scope of Thiamine Diphosphate Dependent Cyclohexane-1,2-Dione Hydrolase: From C-C Bond Cleavage to C-C Bond Ligation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3ZGZ
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
3W5K
 
 | | Crystal structure of Snail1 and importin beta complex | | Descriptor: | Importin subunit beta-1, ZINC ION, Zinc finger protein SNAI1 | | Authors: | Choi, S, Yamashita, E, Yasuhara, N, Song, J, Son, S.Y, Won, Y.H, Shin, Y.S, Sekimoto, T, Park, I.Y, Yoneda, Y, Lee, S.J. | | Deposit date: | 2013-01-30 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the selective nuclear import of the C2H2 zinc-finger protein Snail by importin beta.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3TNF
 
 | | LidA from Legionella in complex with active Rab8a | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LidA, MAGNESIUM ION, ... | | Authors: | Schoebel, S, Cichy, A.L, Goody, R.S, Itzen, A. | | Deposit date: | 2011-09-01 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Protein LidA from Legionella is a Rab GTPase supereffector.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3RMR
 
 | |
3JZA
 
 | | Crystal structure of human Rab1b in complex with the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | PHOSPHATE ION, Ras-related protein Rab-1B, Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3JZ9
 
 | | Crystal structure of the GEF domain of DrrA/SidM from Legionella pneumophila | | Descriptor: | Uncharacterized protein DrrA | | Authors: | Schoebel, S, Oesterlin, L.K, Blankenfeldt, W, Goody, R.S, Itzen, A. | | Deposit date: | 2009-09-23 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity.
Mol.Cell, 36, 2009
|
|
3CAD
 
 | | Crystal structure of Natural Killer Cell Receptor, Ly49G | | Descriptor: | Lectin-related NK cell receptor LY49G1 | | Authors: | Cho, S. | | Deposit date: | 2008-02-19 | | Release date: | 2008-04-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Architecture of the Major Histocompatibility Complex Class I-binding Site of Ly49 Natural Killer Cell Receptors.
J.Biol.Chem., 283, 2008
|
|
5F5B
 
 | |
5F5G
 
 | |
6PJU
 
 | |
6PJR
 
 | |
6PJA
 
 | |
6PJ5
 
 | |
6PJ9
 
 | |
6PJ7
 
 | |
6PJQ
 
 | |
6PJ8
 
 | |
6PJP
 
 | |
6PJ4
 
 | |
5F5J
 
 | |
5F5K
 
 | |
5F5D
 
 | |
3G8K
 
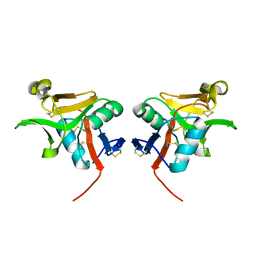 | | Crystal structure of murine natural killer cell receptor, Ly49L4 | | Descriptor: | Lectin-related NK cell receptor LY49L1 | | Authors: | Cho, S. | | Deposit date: | 2009-02-12 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct conformations of Ly49 natural killer cell receptors mediate MHC class I recognition in trans and cis.
Immunity, 31, 2009
|
|
