8WEJ
 
 | |
6BV3
 
 | | Crystal structure of porcine aminopeptidase-N with Leucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
7L7S
 
 | | Human mitochondrial chaperonin mHsp60 | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Chen, L, Wang, J.C.Y. | | Deposit date: | 2020-12-30 | | Release date: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the structural dynamics of human mitochondrial chaperonin mHsp60.
Sci Rep, 11, 2021
|
|
5F9W
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of CH235-lineage antibody CH235, Light chain of CH235-lineage antibody CH235, ... | | Authors: | Chen, L, Zhou, T, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8911 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
5TDG
 
 | |
5TDL
 
 | |
8GZ3
 
 | |
6L48
 
 | |
6L47
 
 | | Structure of the human sterol O-acyltransferase 1 in complex with CI-976 | | Descriptor: | 2,2-dimethyl-N-(2,4,6-trimethoxyphenyl)dodecanamide, CHOLESTEROL, Sterol O-acyltransferase 1 | | Authors: | Chen, L, Guan, C, Niu, Y. | | Deposit date: | 2019-10-16 | | Release date: | 2020-04-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor.
Nat Commun, 11, 2020
|
|
7CU3
 
 | | Structure of mammalian NALCN-FAM155A complex at 2.65 angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Sodium leak channel non-selective protein, ... | | Authors: | Chen, L, Kang, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-18 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of voltage-modulated sodium-selective NALCN-FAM155A channel complex.
Nat Commun, 11, 2020
|
|
3DAE
 
 | |
6JB1
 
 | | Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ATP-binding cassette sub-family C member 8 isoform X2, ATP-sensitive inward rectifier potassium channel 11, ... | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6JPF
 
 | |
6JB3
 
 | | Structure of SUR1 subunit bound with repaglinide | | Descriptor: | ATP-binding cassette sub-family C member 8 isoform X2, Digitonin, Repaglinide | | Authors: | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | Deposit date: | 2019-01-25 | | Release date: | 2019-05-22 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.001 Å) | | Cite: | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6JT2
 
 | | Structure of human soluble guanylate cyclase in the NO activated state | | Descriptor: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | Authors: | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | Deposit date: | 2019-04-08 | | Release date: | 2019-09-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
2LOZ
 
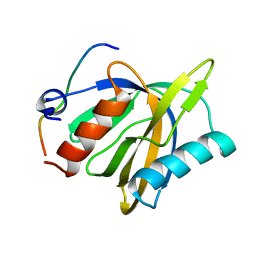 | | The novel binding mode of DLC1 and Tensin2 PTB domain | | Descriptor: | Rho GTPase-activating protein 7, Tensin-like C1 domain-containing phosphatase | | Authors: | Liu, C, Chen, L, Zhu, G. | | Deposit date: | 2012-01-29 | | Release date: | 2012-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the phosphotyrosine binding (PTB) domain of human tensin2 protein in complex with deleted in liver cancer 1 (DLC1) peptide reveals a novel peptide binding mode.
J.Biol.Chem., 287, 2012
|
|
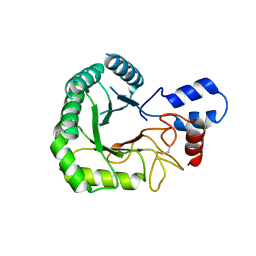
3RJ2
 
 | |
3RPT
 
 | |
3MQ0
 
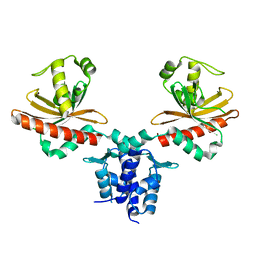 | | Crystal Structure of Agobacterium tumefaciens repressor BlcR | | Descriptor: | Transcriptional repressor of the blcABC operon | | Authors: | Chen, L. | | Deposit date: | 2010-04-27 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | The Agrobacterium tumefaciens Transcription Factor BlcR Is Regulated via Oligomerization.
J.Biol.Chem., 286, 2011
|
|
5ZBG
 
 | |
5YX9
 
 | |
7E3X
 
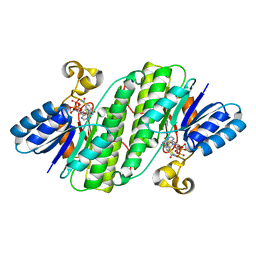 | | Crystal structure of SDR family NAD(P)-dependent oxidoreductase from exiguobacterium | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase | | Authors: | Chen, L, Tang, J, Yuan, S, Zhang, F, Chen, S. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-guided evolution of a ketoreductase forefficient and stereoselective bioreduction of bulkyalpha-aminobeta-keto esters
Catalysis Science And Technology, 2021
|
|
4U1W
 
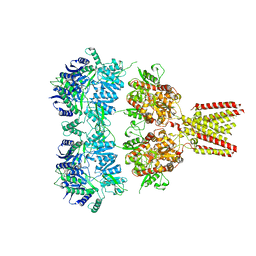 | | Full length GluA2-kainate-(R,R)-2b complex crystal form A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2, ... | | Authors: | Chen, L, Gouaux, E. | | Deposit date: | 2014-07-16 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and Dynamics of AMPA Receptor GluA2 in Resting, Pre-Open, and Desensitized States.
Cell, 158, 2014
|
|
7YSH
 
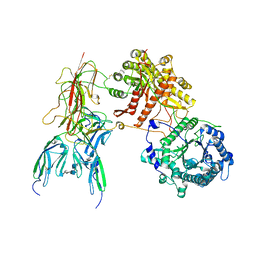 | | Cryo-EM Structure of FGF23-FGFR1c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
7YSU
 
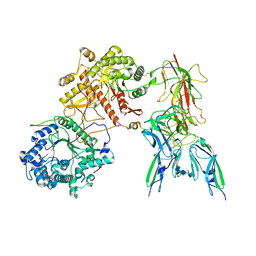 | | Cryo-EM Structure of FGF23-FGFR3c-aKlotho-HS Quaternary Complex | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, COPPER (II) ION, Fibroblast growth factor 23, ... | | Authors: | Mohammadi, M, Chen, L. | | Deposit date: | 2022-08-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for FGF hormone signalling.
Nature, 618, 2023
|
|
