4YCW
 
 | | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by ATP Competitive Inhibitor
Chem.Biol., 22, 2015
|
|
2PWX
 
 | | Crystal structure of G11A mutant of SARS-CoV 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Chen, S, Hu, T, Jiang, H, Shen, X. | | Deposit date: | 2007-05-14 | | Release date: | 2007-10-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutation of Gly-11 on the dimer interface results in the complete crystallographic dimer dissociation of severe acute respiratory syndrome coronavirus 3C-like protease: crystal structure with molecular dynamics simulations.
J.Biol.Chem., 283, 2008
|
|
3QK0
 
 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
4EO7
 
 | | Crystal structure of the TIR domain of human myeloid differentiation primary response protein 88. | | Descriptor: | MAGNESIUM ION, Myeloid differentiation primary response protein MyD88 | | Authors: | Snyder, G.A, Cirl, C, Jiang, J.S, Chen, P, Smith, T, Xiao, T.S. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3QJZ
 
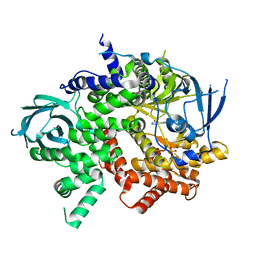 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
3KMW
 
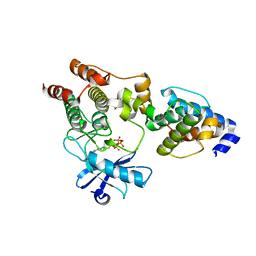 | | Crystal structure of the ILK/alpha-parvin core complex (MgATP) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-parvin, Integrin-linked kinase, ... | | Authors: | Fukuda, K, Qin, J. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The pseudoactive site of ILK is essential for its binding to alpha-Parvin and localization to focal adhesions.
Mol.Cell, 36, 2009
|
|
3KMU
 
 | |
2H1Y
 
 | |
3SHZ
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-chloro-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SHY
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-fluoro-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.647 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
3SIE
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 5-bromo-6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Chen, T.T, Chen, T, Xu, Y.C. | | Deposit date: | 2011-06-17 | | Release date: | 2011-08-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Utilization of Halogen Bond in Lead Optimization: A Case Study of Rational Design of Potent Phosphodiesterase Type 5 (PDE5) Inhibitors.
J.Med.Chem., 54, 2011
|
|
7T6B
 
 | | Structure of S1PR2-heterotrimeric G13 signaling complex | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Li, X, Chen, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-04-06 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structure of S1PR2-heterotrimeric G 13 signaling complex.
Sci Adv, 8, 2022
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8G2O
 
 | | Fab structure - Anti-ApoE-7C11 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, anti-ApoE-7C11 heavy chain, anti-ApoE-7C11 light chain | | Authors: | Marino, C, Arboleda-Velasquez, J.F. | | Deposit date: | 2023-02-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | APOE Christchurch-mimetic therapeutic antibody reduces APOE-mediated toxicity and tau phosphorylation.
Alzheimers Dement, 20, 2024
|
|
3RSV
 
 | |
3RTH
 
 | |
3RTN
 
 | |
5B73
 
 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
7VD4
 
 | | Crystal structure of BPTF-BRD with ligand TP248 bound | | Descriptor: | 6-[4-[3-(dimethylamino)propoxy]phenyl]-N-methyl-2-methylsulfonyl-pyrimidin-4-amine, Nucleosome-remodeling factor subunit BPTF | | Authors: | Lu, T, Lu, H.B. | | Deposit date: | 2021-09-06 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85659146 Å) | | Cite: | Discovery of a highly potent CECR2 bromodomain inhibitor with 7H-pyrrolo[2,3-d] pyrimidine scaffold.
Bioorg.Chem., 123, 2022
|
|
2BX4
 
 | |
2BX3
 
 | |
2GVF
 
 | | HCV NS3-4A protease domain complexed with a macrocyclic ketoamide inhibitor, SCH419021 | | Descriptor: | (6R,8S,11S)-11-CYCLOHEXYL-N-(1-{[(2-{[(1S)-2-(DIMETHYLAMINO)-2-OXO-1-PHENYLETHYL]AMINO}-2-OXOETHYL)AMINO](OXO)ACETYL}BUTYL)-10,13-DIOXO-2,5-DIOXA-9,12-DIAZATRICYCLO[14.3.1.1~6,9~]HENICOSA-1(20),16,18-TRIENE-8-CARBOXAMIDE, Polyprotein, ZINC ION, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chen, K.X, Venkatraman, S, Parekh, T.N, Gu, H, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V, Girijavallabhan, V. | | Deposit date: | 2006-05-02 | | Release date: | 2007-01-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7EPU
 
 | | Crystal structure of HsALC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromodomain-helicase-DNA-binding protein 1-like, MAGNESIUM ION, ... | | Authors: | Wang, L, Chen, K.J. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
6VB9
 
 | | Covalent adduct of cis-2,3-epoxysuccinic acid with Isocitrate Lyase-1 from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Isocitrate lyase, ... | | Authors: | Krieger, I.V, Pham, T.V, Meek, T.D, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2019-12-18 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Covalent Inactivation of Mycobacterium tuberculosis Isocitrate Lyase by cis -2,3-Epoxy-Succinic Acid.
Acs Chem.Biol., 16, 2021
|
|
4RLV
 
 | |
