3R0W
 
 | | Crystal Structures of Multidrug-resistant HIV-1 Protease in Complex with Mechanism-Based Aspartyl Protease Inhibitors. | | Descriptor: | Multidrug-resistant clinical isolate 769 HIV-1 Protease, N-[(2R)-1-{[(2S,3S)-5-{[(2R)-1-{[(2S)-1-amino-4-methyl-1-oxopentan-2-yl]amino}-3-chloro-1-oxopropan-2-yl]amino}-3-hydroxy-5-oxo-1-phenylpentan-2-yl]amino}-3-methyl-1-oxobutan-2-yl]pyridine-2-carboxamide | | Authors: | Yedidi, R.S, Gupta, D, Liu, Z, Brunzelle, J, Kovari, I.A, Woster, P.M, Kovari, L.C. | | Deposit date: | 2011-03-09 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of multidrug-resistant HIV-1 protease in complex with two potent anti-malarial compounds.
Biochem.Biophys.Res.Commun., 421, 2012
|
|
2R31
 
 | | Crystal structure of atp12p from paracoccus denitrificans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ATP12 ATPase | | Authors: | Ludlam, A.V, Brunzelle, J.S, Gatti, D.L, Ackerman, S.H. | | Deposit date: | 2007-08-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chaperones of F1-ATPase.
J.Biol.Chem., 284, 2009
|
|
5TO5
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO6
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Bandyopadhyay, A, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TO7
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-10-16 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5TVB
 
 | | Structure of the TPR oligomerization domain | | Descriptor: | Nucleoprotein TPR | | Authors: | Pal, K, Xu, Q, Zhou, X.E, Melcher, K, Xu, H.E. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Structure, 25, 2017
|
|
5VBC
 
 | | Crystal structure of ATXR5 in complex with histone H3.1 | | Descriptor: | DIMETHYL SULFOXIDE, Histone H3.1 peptide, Probable Histone-lysine N-methyltransferase ATXR5, ... | | Authors: | Couture, J.-F, Bergamin, E. | | Deposit date: | 2017-03-29 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the methylation specificity of ATXR5 for histone H3.
Nucleic Acids Res., 45, 2017
|
|
5IZQ
 
 | | Crystal structure of human folate receptor alpha in complex with novel antifolate AGF183 | | Descriptor: | Folate receptor alpha, N-(4-{[2-(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-6-yl)ethyl]amino}benzene-1-carbonyl)-L-glutamic acid | | Authors: | Ke, J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2016-03-25 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Tumor Targeting with Novel 6-Substituted Pyrrolo [2,3-d] Pyrimidine Antifolates with Heteroatom Bridge Substitutions via Cellular Uptake by Folate Receptor alpha and the Proton-Coupled Folate Transporter and Inhibition of de Novo Purine Nucleotide Biosynthesis.
J.Med.Chem., 59, 2016
|
|
4O30
 
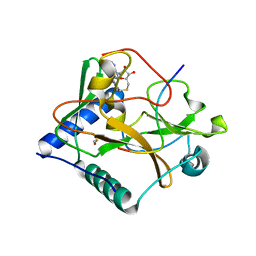 | | Crystal structure of ATXR5 in complex with histone H3.1 and AdoHcy | | Descriptor: | BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, Histone-lysine N-methyltransferase ATXR6, ... | | Authors: | Bergamin, E, Mongeon, V, Couture, J.F. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication.
Science, 343, 2014
|
|
2Q12
 
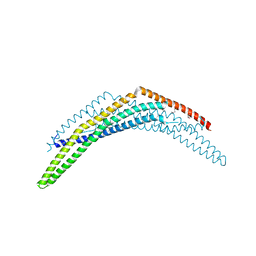 | | Crystal Structure of BAR domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhang, X.C, Zhu, G. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
2Q13
 
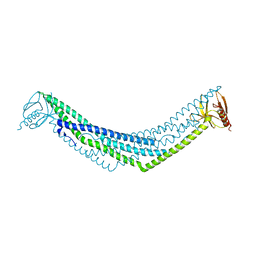 | | Crystal structure of BAR-PH domain of APPL1 | | Descriptor: | DCC-interacting protein 13 alpha | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2007-05-23 | | Release date: | 2007-08-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the APPL1 BAR-PH domain and characterization of its interaction with Rab5.
Embo J., 26, 2007
|
|
3ERP
 
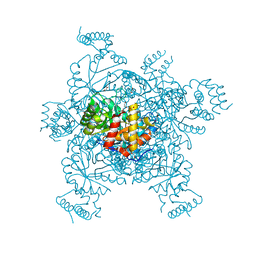 | | Structure of IDP01002, a putative oxidoreductase from and essential gene of Salmonella typhimurium | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Minasov, G, Evdokimova, E, Brunzelle, J.S, Kudritska, M, Edwards, A.M, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-10-02 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl.Environ.Microbiol., 2017
|
|
4YO4
 
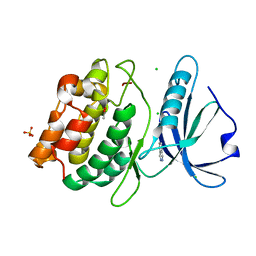 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment phthalazine | | Descriptor: | ACETATE ION, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
4YPD
 
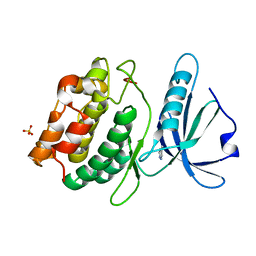 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
3D6N
 
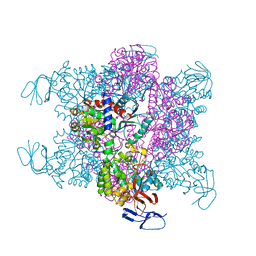 | | Crystal Structure of Aquifex Dihydroorotase Activated by Aspartate Transcarbamoylase | | Descriptor: | Aspartate carbamoyltransferase, CITRATE ANION, Dihydroorotase, ... | | Authors: | Edwards, B.F.P. | | Deposit date: | 2008-05-20 | | Release date: | 2009-01-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dihydroorotase from the hyperthermophile Aquifiex aeolicus is activated by stoichiometric association with aspartate transcarbamoylase and forms a one-pot reactor for pyrimidine biosynthesis.
Biochemistry, 48, 2009
|
|
3F8T
 
 | |
5EJN
 
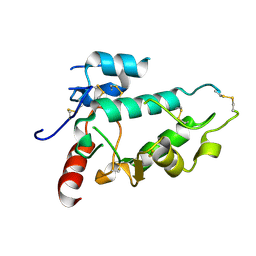 | |
5T0Q
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 Jas domain [166-192] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T0F
 
 | | Crystal structure of the Myc3 N-terminal domain [44-242] in complex with JAZ10 CMID domain [16-58] from arabidopsis | | Descriptor: | Protein TIFY 9, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Brunzelle, J.S, He, S.Y, Xu, H.E, Melcher, K. | | Deposit date: | 2016-08-16 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into alternative splicing-mediated desensitization of jasmonate signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UWG
 
 | | The crystal structure of Fz4-CRD in complex with palmitoleic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, PALMITOLEIC ACID | | Authors: | Ke, J, DeBruine, Z.J, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2017-02-21 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Wnt5a promotes Frizzled-4 signalosome assembly by stabilizing cysteine-rich domain dimerization.
Genes Dev., 31, 2017
|
|
4LEU
 
 | | Crystal Structure of THA8-like protein from Arabidopsis thaliana | | Descriptor: | Pentatricopeptide repeat-containing protein At3g46870 | | Authors: | Ke, J, Chen, R.Z, Ban, T, Brunzelle, J.S, Gu, X, Kang, Y, Melcher, K, Zhu, J.K, Xu, H.E. | | Deposit date: | 2013-06-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a PLS-class Pentatricopeptide Repeat Protein Provides Insights into Mechanism of RNA Recognition.
J.Biol.Chem., 288, 2013
|
|
3K29
 
 | | Structure of a putative YscO homolog CT670 from Chlamydia trachomatis | | Descriptor: | Putative uncharacterized protein | | Authors: | Lam, R, Singer, A, Skarina, T, Onopriyenko, O, Bochkarev, A, Brunzelle, J.S, Edwards, A.M, Anderson, W.F, Chirgadze, N.Y, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-13 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and protein-protein interaction studies on Chlamydia trachomatis protein CT670 (YscO Homolog).
J.Bacteriol., 192, 2010
|
|
