5N53
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with N-(3-chloro-4-methoxyphenyl) acetamide | | Descriptor: | D-3-phosphoglycerate dehydrogenase, ~{N}-(3-chloranyl-4-methoxy-phenyl)ethanamide | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-12 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5N6C
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with NAD and L-Tartrate | | Descriptor: | D-3-phosphoglycerate dehydrogenase, L(+)-TARTARIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Unterlass, J.E, Basle, A, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-02-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the enzymatic activity and potential substrate promiscuity of human 3-phosphoglycerate dehydrogenase (PHGDH).
Oncotarget, 8, 2017
|
|
5NUP
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ABC transporter permease, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NUQ
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein F, Probable phospholipid-binding lipoprotein mlaA | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NZQ
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-(1,3-oxazol-5-yl)aniline. | | Descriptor: | 3-(1,3-oxazol-5-yl)aniline, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NUR
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(3-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NUO
 
 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, ABC transporter permease, Outer membrane protein F, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
5NZP
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Hydroxybenzisoxazole | | Descriptor: | 1,2-benzoxazol-3-ol, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFV
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-fluoro-2-methylbenzoic acid | | Descriptor: | 5-fluoranyl-2-methyl-benzoic acid, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5NZO
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 1-methyl-3-phenyl-1H-pyrazol-5-amine | | Descriptor: | 2-methyl-5-phenyl-pyrazol-3-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-05-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFW
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 3-Chloro-4-fluorobenzamide | | Descriptor: | 3-chloranyl-4-fluoranyl-benzamide, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Validating and enabling phosphoglycerate dehydrogenase (PHGDH) as a target for fragment-based drug discovery in PHGDH-amplified breast cancer.
Oncotarget, 9, 2018
|
|
5OFM
 
 | | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole | | Descriptor: | 1-methylindol-5-amine, D-3-phosphoglycerate dehydrogenase | | Authors: | Unterlass, J.E, Basle, A, Blackburn, T.J, Tucker, J, Cano, C, Noble, M.E.M, Curtin, N.J. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human 3-phosphoglycerate dehydrogenase in complex with 5-amino-1-methyl-1H-indole
To be published
|
|
4V1L
 
 | | High resolution structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V1K
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 9 (Cel9A) from Ruminococcus flavefaciens FD-1 | | Descriptor: | 2-HYDROXY BUTANE-1,4-DIOL, CALCIUM ION, CARBOHYDRATE BINDING MODULE, ... | | Authors: | Venditto, I, Goyal, A, Thompson, A, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-29 | | Release date: | 2016-01-20 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V1I
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 at medium resolution | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-26 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V18
 
 | | SeMet structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V17
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4V1B
 
 | | Structure of a novel carbohydrate binding module from glycoside hydrolase family 5 glucanase from Ruminococcus flavefaciens FD-1 collected at the Zn edge | | Descriptor: | CARBOHYDRATE BINDING MODULE | | Authors: | Venditto, I, Centeno, M.S.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2014-09-25 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Complexity of the Ruminococcus Flavefaciens Cellulosome Reflects an Expansion in Glycan Recognition.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5AFE
 
 | |
2BAS
 
 | | Crystal Structure of the Bacillus subtilis YkuI Protein, with an EAL Domain. | | Descriptor: | BETA-MERCAPTOETHANOL, YkuI protein | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Miller, D.J, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-10-14 | | Release date: | 2005-11-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structures of YkuI and its complex with second messenger cyclic Di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains.
J.Biol.Chem., 284, 2009
|
|
5G56
 
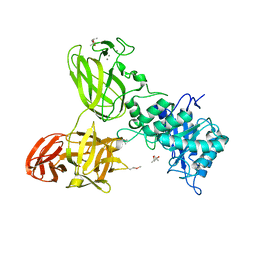 | | THE TETRA-MODULAR CELLULOSOMAL ARABINOXYLANASE CtXyl5A STRUCTURE AS REVEALED BY X-RAY CRYSTALLOGRAPHY | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, CARBOHYDRATE BINDING FAMILY 6 | | Authors: | Bras, J.L.A, Gilbert, H.J, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2016-05-21 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Mechanism by which Arabinoxylanases Can Recognise Highly Decorated Xylans.
J.Biol.Chem., 291, 2016
|
|
5T87
 
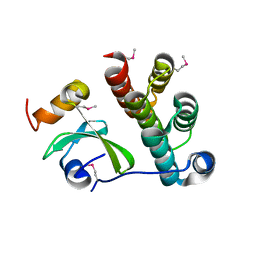 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
5AEZ
 
 | |
5AEX
 
 | |
5AID
 
 | |
