5D4U
 
 | | SAM-bound HcgC from Methanocaldococcus jannaschii | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, Uncharacterized protein MJ0489 | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
6DL2
 
 | | BRD4 bromodomain 1 in complex with HYB157 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-2,9-dimethyl-4H,6H-thieno[2,3-e][1,2,4]triazolo[3,4-c][1,4]oxazepine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-05-31 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery of QCA570 as an Exceptionally Potent and Efficacious Proteolysis Targeting Chimera (PROTAC) Degrader of the Bromodomain and Extra-Terminal (BET) Proteins Capable of Inducing Complete and Durable Tumor Regression.
J. Med. Chem., 61, 2018
|
|
7F70
 
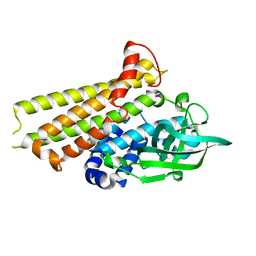 | | Crystal structure of Rv3094c | | Descriptor: | Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y, Zhang, H.T. | | Deposit date: | 2021-06-26 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F74
 
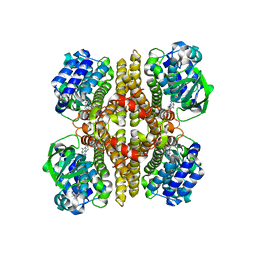 | | Rv3094c in complex with FMN. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
7F72
 
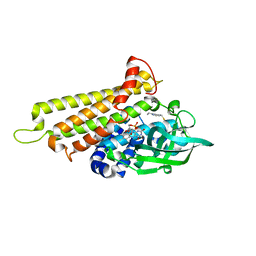 | | Rv3094c in complex with FAD and ETH. | | Descriptor: | 2-ethylpyridine-4-carboximidothioic acid, FLAVIN MONONUCLEOTIDE, Rv3094c | | Authors: | Wang, Z.X, Ouyang, S.Y. | | Deposit date: | 2021-06-27 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Omics analysis of Mycobacterium tuberculosis isolates uncovers Rv3094c, an ethionamide metabolism-associated gene.
Commun Biol, 6, 2023
|
|
8Y33
 
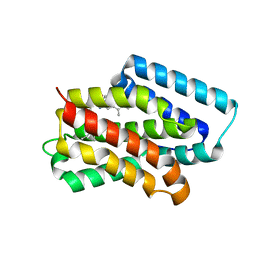 | | A near-infrared fluorescent protein of de novo backbone design | | Descriptor: | 3-[5-[(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, near-infrared fluorescent protein | | Authors: | Hu, X, Xu, Y. | | Deposit date: | 2024-01-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Using Protein Design and Directed Evolution to Monomerize a Bright Near-Infrared Fluorescent Protein.
Acs Synth Biol, 13, 2024
|
|
3DPC
 
 | |
4Z93
 
 | | BRD4 bromodomain 2 in complex with gamma-carboline-containing compound, number 18. | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-cyclopropyl-5-methyl-1H-pyrazol-4-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-8-methoxy-5H-pyrido[4,3-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2015-04-09 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structure-Based Design of gamma-Carboline Analogues as Potent and Specific BET Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5TRF
 
 | | MDM2 in complex with SAR405838 | | Descriptor: | (2'S,3R,4'S,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(trans-4-hydroxycyclohexyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.
Cancer Res., 74, 2014
|
|
5KWA
 
 | |
7S05
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S06
 
 | | Cryo-EM structure of human GlcNAc-1-phosphotransferase A2B2 subcomplex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosamine-1-phosphotransferase subunits alpha/beta | | Authors: | Li, H, Li, H. | | Deposit date: | 2021-08-30 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the human GlcNAc-1-phosphotransferase alpha beta subunits reveals regulatory mechanism for lysosomal enzyme glycan phosphorylation.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5UOO
 
 | | BRD4 bromodomain 2 in complex with CD161 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-17 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Discovery of 4-(6-Methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethylisoxazole (CD161) as a Potent and Orally Bioavailable BET Bromodomain Inhibitor.
J. Med. Chem., 60, 2017
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
8HN9
 
 | | Human SIRT3 Recognizing CCNE2K348la peptide | | Descriptor: | CCNE2 peptide, IMIDAZOLE, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Wang, Y, Ding, W. | | Deposit date: | 2022-12-07 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | SIRT3-dependent delactylation of cyclin E2 prevents hepatocellular carcinoma growth.
Embo Rep., 24, 2023
|
|
8J1X
 
 | |
8J1W
 
 | |
5D4T
 
 | |
5D4V
 
 | | HcgC with SAH and a guanylylpyridinol (GP) derivative | | Descriptor: | 5'-O-[(R)-[(3,6-dimethyl-2-oxo-1,2-dihydropyridin-4-yl)oxy](hydroxy)phosphoryl]guanosine, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of HcgC as a SAM-Dependent Pyridinol Methyltransferase in [Fe]-Hydrogenase Cofactor Biosynthesis.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5D5T
 
 | |
3SP7
 
 | | Crystal Structure of Bcl-xL bound to BM903 | | Descriptor: | 5-(4-chlorophenyl)-4-{3-[4-(4-{[(4-{[(2R)-4-(dimethylamino)-1-(phenylsulfanyl)butan-2-yl]amino}-3-nitrophenyl)sulfonyl]amino}phenyl)piperazin-1-yl]phenyl}-1,2-dimethyl-1H-pyrrole-3-carboxylic acid, ACETATE ION, Bcl-2-like protein 1, ... | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-07-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-based design of a new class of potent Bcl-2/Bcl-xL inhibitors
To be Published
|
|
3SPF
 
 | | Crystal Structure of Bcl-xL bound to BM501 | | Descriptor: | 4-(4-chlorophenyl)-1-[(3S)-3,4-dihydroxybutyl]-N-[3-(4-methylpiperazin-1-yl)propyl]-3-phenyl-1H-pyrrole-2-carboxamide, Bcl-2-like protein 1, GLYCEROL | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2011-07-01 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Bcl-2 and Bcl-xL Inhibitors with Subnanomolar Binding Affinities Based upon a New Scaffold.
J.Med.Chem., 55, 2012
|
|
5ZB1
 
 | | Monomeric crystal structure of orf57 from KSHV | | Descriptor: | ORF57, ZINC ION | | Authors: | Gao, Z.Q, Yuan, F, Lan, K, Dong, Y.H. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.061 Å) | | Cite: | The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.
PLoS Pathog., 14, 2018
|
|
5ZB3
 
 | | Dimeric crystal structure of ORF57 from KSHV | | Descriptor: | ORF57, ZINC ION | | Authors: | Gao, Z.Q, Yuan, F, Dong, Y.H, Lan, K. | | Deposit date: | 2018-02-09 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.506 Å) | | Cite: | The crystal structure of KSHV ORF57 reveals dimeric active sites important for protein stability and function.
PLoS Pathog., 14, 2018
|
|
