2N92
 
 | | Solution structure of cecropin P1 with LPS | | Descriptor: | Cecropin-P1 | | Authors: | Baek, M, Kamiya, M, Kushibiki, T, Nakazumi, T, Tomisawa, S, Abe, C, Kumaki, Y, Kikukawa, T, Demura, M, Kawano, K, Aizawa, T. | | Deposit date: | 2015-11-04 | | Release date: | 2016-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Lipopolysaccharide bound structure of antimicrobial peptide cecropin P1 by NMR spectroscopy
To be Published
|
|
8FNE
 
 | | phiPA3 PhuN Tetramer, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2022-12-27 | | Release date: | 2023-03-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8VC8
 
 | |
8SK7
 
 | | Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA_20 minibinder (RFdiffusion-designed), ... | | Authors: | Borst, A.J, Bennett, N.R. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | De novo design of protein structure and function with RFdiffusion.
Nature, 620, 2023
|
|
7MEZ
 
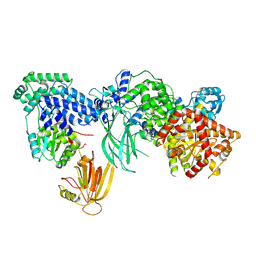 | | Structure of the phosphoinositide 3-kinase p110 gamma (PIK3CG) p101 (PIK3R5) complex | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, Phosphoinositide 3-kinase regulatory subunit 5 | | Authors: | Burke, J.E, Dalwadi, U, Rathinaswamy, M.K, Yip, C.K. | | Deposit date: | 2021-04-08 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of the phosphoinositide 3-kinase (PI3K) p110 gamma-p101 complex reveals molecular mechanism of GPCR activation.
Sci Adv, 7, 2021
|
|
7QFJ
 
 | | Crystal structure of S-layer protein SlpX from Lactobacillus acidophilus, domain II (aa 194-362) | | Descriptor: | SlpX | | Authors: | Sagmeister, T, Pavkov-Keller, T, Buhlheller, C, Baek, M, Read, R, Baker, D. | | Deposit date: | 2021-12-06 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The molecular architecture of Lactobacillus S-layer: Assembly and attachment to teichoic acids.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5ZWV
 
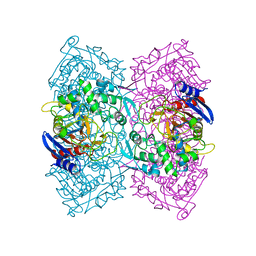 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29 | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W. | | Deposit date: | 2018-05-17 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWR
 
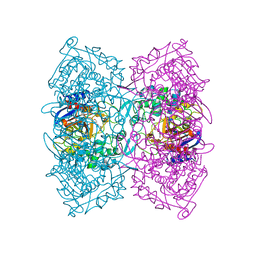 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | (2S)-2-[3-(benzenecarbonyl)phenyl]propanoic acid, Est-Y29, GLYCEROL | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
5ZWQ
 
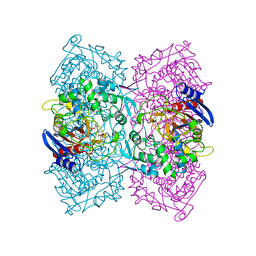 | | Structural Basis for the Enantioselectivity of Est-Y29 toward (S)-ketoprofen | | Descriptor: | Est-Y29, GLYCEROL, ethyl (2S)-2-[3-(benzenecarbonyl)phenyl]propanoate | | Authors: | Ngo, D.T, Oh, C, Park, K, Nguyen, L, Byun, H.M, Kim, S, Yoon, S, Ryu, Y, Ryu, B.H, Kim, T.D, Yang, J.W, Kim, K.K. | | Deposit date: | 2018-05-16 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural Basis for the Enantioselectivity of Esterase Est-Y29 toward (S)-Ketoprofen
Acs Catalysis, 9, 2019
|
|
6JKG
 
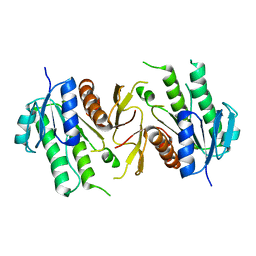 | | The NAD+-free form of human NSDHL | | Descriptor: | Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
6JKH
 
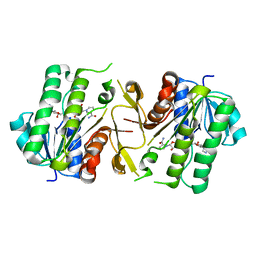 | | The NAD+-bound form of human NSDHL | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Sterol-4-alpha-carboxylate 3-dehydrogenase, decarboxylating | | Authors: | Kim, D, Lee, S.J, Lee, B. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of human NSDHL and development of its novel inhibitor with the potential to suppress EGFR activity.
Cell.Mol.Life Sci., 78, 2021
|
|
7BOT
 
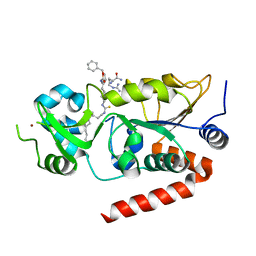 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.23 | | Descriptor: | N-dodecylmethanethioamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
7BOS
 
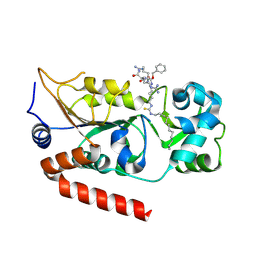 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.13 | | Descriptor: | Myristoyl thiourea inhibitor, No.13, N-dodecylmethanethioamide, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
8DT0
 
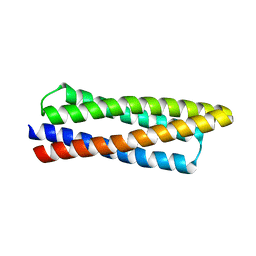 | |
8F54
 
 | |
8F4X
 
 | |
8F53
 
 | |
8D04
 
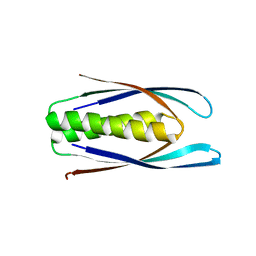 | | Hallucinated C2 protein assembly HALC2_062 | | Descriptor: | HALC2_062 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D07
 
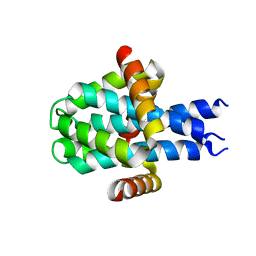 | | Hallucinated C3 protein assembly HALC3_109 | | Descriptor: | HALC3_109 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D05
 
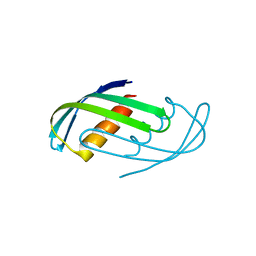 | | Hallucinated C2 protein assembly HALC2_065 | | Descriptor: | HALC2_065 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D08
 
 | | Hallucinated C4 protein assembly HALC4_135 | | Descriptor: | HALC4_135 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D03
 
 | | Hallucinated C2 protein assembly HALC2_068 | | Descriptor: | HALC2_068 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D09
 
 | | Hallucinated C4 protein assembly HALC4_136 | | Descriptor: | HALC4_136 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
8D06
 
 | | Hallucinated C3 protein assembly HALC3_104 | | Descriptor: | HALC3_104 | | Authors: | Ragotte, R.J, Bera, A.K, Wicky, B.I.M, Milles, L.F, Baker, D. | | Deposit date: | 2022-05-25 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Hallucinating symmetric protein assemblies.
Science, 378, 2022
|
|
