8W77
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c only | | Descriptor: | Human Consensus Olfactory Receptor OR52c in apo state, receptor only,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8HTI
 
 | | Human Consensus Olfactory Receptor OR52c in Complex with Octanoic Acid (OCA) and G Protein | | Descriptor: | Consensus Olfactory Receptor OR52c, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
8J46
 
 | | Human Consensus Olfactory Receptor OR52c in apo state, OR52c-bRIL | | Descriptor: | Olfactory receptor OR52c,Soluble cytochrome b562 | | Authors: | Choi, C.W, Bae, J, Choi, H.-J, Kim, J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
1OXD
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
6YI9
 
 | | Crystal structure of the rat cytosolic PCK1, acetylated on Lys244 | | Descriptor: | 1,2-ETHANEDIOL, Phosphoenolpyruvate carboxykinase, cytosolic [GTP] | | Authors: | Latorre-Muro, P, Baeza, J, Hurtado-Guerrero, R, Hicks, T, Delso, I, Hernandez-Ruiz, C, Velazquez-Campoy, A, Lawton, A.J, Angulo, J, Denu, J.M, Carrodeguas, J.A. | | Deposit date: | 2020-04-01 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Self-acetylation at the active site of phosphoenolpyruvate carboxykinase (PCK1) controls enzyme activity.
J.Biol.Chem., 296, 2021
|
|
2J8P
 
 | | NMR structure of C-terminal domain of human CstF-64 | | Descriptor: | CLEAVAGE STIMULATION FACTOR 64 KDA SUBUNIT | | Authors: | Qu, X, Perez-Canadillas, J.M, Agrawal, S, De Baecke, J, Cheng, H, Varani, G, Moore, C. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-Terminal Domains of Vertebrate Cstf-64 and its Yeast Orthologue RNA15 Form a New Structure Critical for Mrna 3'-End Processing.
J.Biol.Chem., 282, 2007
|
|
7YON
 
 | | Complex structure of Neuropeptide Y Y2 receptor in complex with PYY(3-36) and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kang, H, Park, C, Kim, J, Choi, H.-J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
7YOO
 
 | | Complex structure of Neuropeptide Y Y2 receptor in complex with NPY and Gi | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Kang, H, Park, C, Kim, J, Choi, H.-J. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for Y2 receptor-mediated neuropeptide Y and peptide YY signaling.
Structure, 31, 2023
|
|
8HTG
 
 | | Crystal structure of Golf in complex with GTP-gamma S and Mg | | Descriptor: | 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Guanine nucleotide-binding protein G(olf) subunit alpha, ... | | Authors: | Kang, H, Choi, H.-J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Understanding the molecular mechanisms of odorant binding and activation of the human OR52 family.
Nat Commun, 14, 2023
|
|
7E72
 
 | | Crystal structure of Tie2-agonistic antibody in complex with human Tie2 Fn2-3 | | Descriptor: | 1,2-ETHANEDIOL, Angiopoietin-1 receptor, the chimeric Fab fragment of 3H7 (heavy chain), ... | | Authors: | Kim, H.M, Jo, G.H, Hong, H.J, Han, A. | | Deposit date: | 2021-02-25 | | Release date: | 2021-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Structural insights into the clustering and activation of Tie2 receptor mediated by Tie2 agonistic antibody.
Nat Commun, 12, 2021
|
|
2FJK
 
 | | Crystal structure of Fructose-1,6-Bisphosphate Aldolase in Thermus caldophilus | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase | | Authors: | Lee, J.H, Im, Y.J, Rho, S.-H, Kim, M.-K, Kang, G.B, Eom, S.H. | | Deposit date: | 2006-01-03 | | Release date: | 2006-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stereoselectivity of fructose-1,6-bisphosphate aldolase in Thermus caldophilus
Biochem.Biophys.Res.Commun., 347, 2006
|
|
2IE8
 
 | |
7VOV
 
 | | The crystal structure of human forkhead box protein in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOX
 
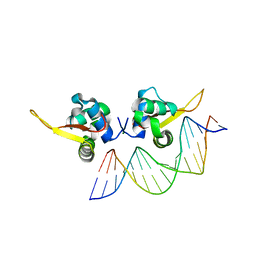 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
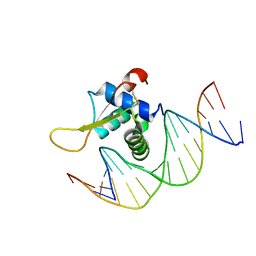 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7QL5
 
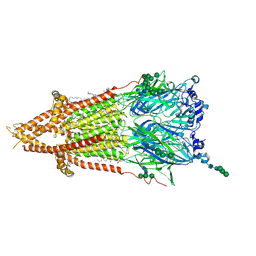 | | Torpedo muscle-type nicotinic acetylcholine receptor - nicotine-bound conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (S)-3-(1-METHYLPYRROLIDIN-2-YL)PYRIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-19 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
7QKO
 
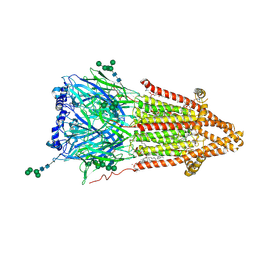 | | Torpedo muscle-type nicotinic acetylcholine receptor - Resting conformation | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Acetylcholine receptor subunit alpha, Acetylcholine receptor subunit beta, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-18 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
7QL6
 
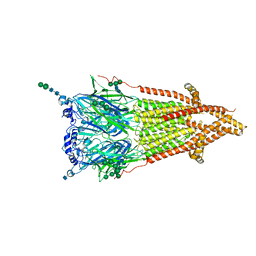 | | Torpedo muscle-type nicotinic acetylcholine receptor - carbamylcholine-bound conformation | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zarkadas, E, Pebay-Peyroula, E, Baenziger, J, Nury, H. | | Deposit date: | 2021-12-19 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Conformational transitions and ligand-binding to a muscle-type nicotinic acetylcholine receptor.
Neuron, 110, 2022
|
|
4U3E
 
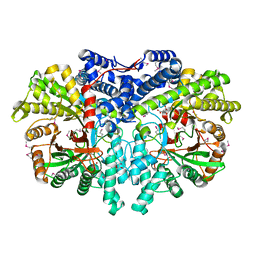 | | Anaerobic ribonucleotide reductase | | Descriptor: | ACETATE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Funk, M.A, Drennan, C.L. | | Deposit date: | 2014-07-20 | | Release date: | 2014-09-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The class III ribonucleotide reductase from Neisseria bacilliformis can utilize thioredoxin as a reductant.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8FJN
 
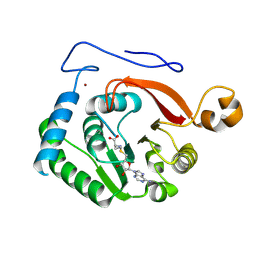 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - C2221 space group | | Descriptor: | CALCIUM ION, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
8FJM
 
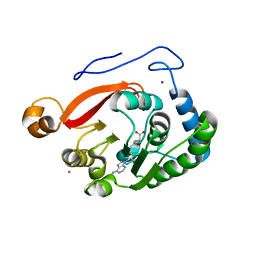 | | Crystal Structure of the Trypanosoma brucei DOT1A histone H3K76 methyltransferase in complex with AdoHcy - P212121 space group | | Descriptor: | ACETATE ION, CALCIUM ION, Histone-lysine N-methyltransferase, ... | | Authors: | Frisbie, V.S, Hashimoto, H, Debler, E.W. | | Deposit date: | 2022-12-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two DOT1 enzymes cooperatively mediate efficient ubiquitin-independent histone H3 lysine 76 tri-methylation in kinetoplastids.
Nat Commun, 15, 2024
|
|
6O76
 
 | | Human cytosolic Histidyl-tRNA synthetase (HisRS) with WHEP domain | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic | | Authors: | Kuhle, B, Yang, X.L. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6APL
 
 | | Crystal Structure of human ST6GALNAC2 in complex with CMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Forouhar, F, Moremen, K.W, Northeast Structural Genomics Consortium (NESG), Tong, L. | | Deposit date: | 2017-08-17 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Expression system for structural and functional studies of human glycosylation enzymes.
Nat. Chem. Biol., 14, 2018
|
|
