2OW3
 
 | | Glycogen synthase kinase-3 beta in complex with bis-(indole)maleimide pyridinophane inhibitor | | Descriptor: | BIS-(INDOLE)MALEIMIDE PYRIDINOPHANE, Glycogen synthase kinase-3 beta | | Authors: | Zhang, H.C, Bonaga, L.V, Ye, H, Derian, C.K, Damiano, B.P, Maryanoff, B.E. | | Deposit date: | 2007-02-15 | | Release date: | 2008-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel bis(indolyl)maleimide pyridinophanes that are potent, selective inhibitors of glycogen synthase kinase-3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5AYR
 
 | | The crystal structure of SAUGI/human UDG complex | | Descriptor: | MAGNESIUM ION, Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
5AYS
 
 | | Crystal structure of SAUGI/HSV UDG complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Huang, M.F, Wang, A.H.J. | | Deposit date: | 2015-09-02 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Using structural-based protein engineering to modulate the differential inhibition effects of SAUGI on human and HSV uracil DNA glycosylase.
Nucleic Acids Res., 44, 2016
|
|
3WDG
 
 | | Staphylococcus aureus UDG / UGI complex | | Descriptor: | Uncharacterized protein, Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
3WDF
 
 | | Staphylococcus aureus UDG | | Descriptor: | Uracil-DNA glycosylase | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2013-06-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Staphylococcus aureus protein SAUGI acts as a uracil-DNA glycosylase inhibitor.
Nucleic Acids Res., 42, 2013
|
|
8J8Y
 
 | |
3W1O
 
 | | Neisseria DNA mimic protein DMP12 | | Descriptor: | DNA mimic protein DMP12, MAGNESIUM ION | | Authors: | Wang, H.C, Ko, T.P, Wu, M.L, Wang, A.H.J. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Neisseria conserved hypothetical protein DMP12 is a DNA mimic that binds to histone-like HU protein
Nucleic Acids Res., 41, 2013
|
|
2ATP
 
 | | Crystal structure of a CD8ab heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD8 alpha chain, T-cell surface glycoprotein CD8 beta chain, ... | | Authors: | Chang, H.C, Tan, K, Ouyang, J, Parisini, E, Liu, J.H, Le, Y, Wang, X, Reinherz, E.L, Wang, J.H. | | Deposit date: | 2005-08-25 | | Release date: | 2005-12-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mutational Analyses of a CD8alphabeta Heterodimer and Comparison with the CD8alphaalpha Homodimer.
Immunity, 23, 2005
|
|
3X0T
 
 | | Crystal structure of PirA | | Descriptor: | NITRATE ION, Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6A9S
 
 | | The crystal structure of vaccinia virus A26 (residues 1-397) | | Descriptor: | 1,2-ETHANEDIOL, Protein A26 | | Authors: | Wang, H.C, Ko, T.Z, Luo, Y.C, Liao, Y.T, Chang, W. | | Deposit date: | 2018-07-16 | | Release date: | 2019-06-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Vaccinia viral A26 protein is a fusion suppressor of mature virus and triggers membrane fusion through conformational change at low pH.
Plos Pathog., 15, 2019
|
|
3X0U
 
 | | Crystal structure of PirB | | Descriptor: | Uncharacterized protein | | Authors: | Wang, H.C, Ko, T.P, Wang, A.H.J, Lo, C.F. | | Deposit date: | 2014-10-22 | | Release date: | 2015-08-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5JX5
 
 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2016-05-12 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo- and endo- activity in a complex with cellobiose
Acta Crystallogr.,Sect.D, 2019
|
|
6IDW
 
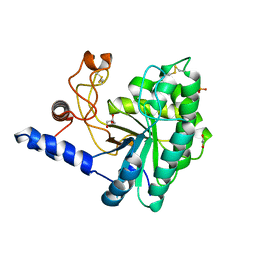 | | GH6 Orpinomyces sp. Y102 enzyme | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Tsai, L.C, Huang, H.C. | | Deposit date: | 2018-09-11 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structures of the GH6 Orpinomyces sp. Y102 CelC7 enzyme with exo and endo activity and its complex with cellobiose.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8TQC
 
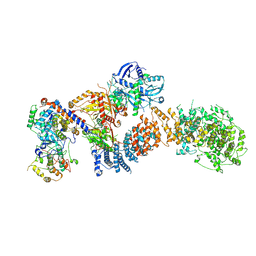 | | Structure of the human CDK8 kinase module | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 12, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-06 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TQ2
 
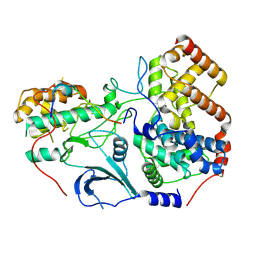 | | Structure of the kinase lobe of human CDK8 kinase module | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 12, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-06 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TRH
 
 | | The IDRc bound human core Mediator complex | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
8TQW
 
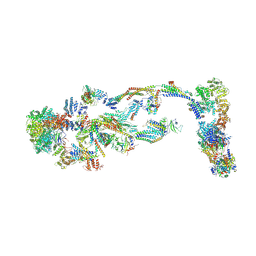 | | Structure of human transcriptional Mediator complex | | Descriptor: | Cyclin-C, Cyclin-dependent kinase 8, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Chen, S.F, Chao, T.C, Kim, H.J, Tang, H.C, Khadka, S, Li, T, Murakami, K, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2023-08-08 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of the human transcriptional Mediator regulated by its dissociable kinase module.
Mol.Cell, 84, 2024
|
|
2J20
 
 | |
3FHA
 
 | | Structure of endo-beta-N-acetylglucosaminidase A | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase, GLYCEROL, ... | | Authors: | Yin, J, Li, L, Shaw, N, Li, Y, Song, J.K, Zhang, W, Xia, C, Zhang, R, Joachimiak, A, Zhang, H.C, Wang, L.X, Wang, P, Liu, Z.J. | | Deposit date: | 2008-12-09 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis and catalytic mechanism for the dual functional endo-beta-N-acetylglucosaminidase A.
Plos One, 4, 2009
|
|
8INI
 
 | | vaccinia H2 protein without the transmembrane region | | Descriptor: | TETRAETHYLENE GLYCOL, vaccinia h2 protein | | Authors: | Liu, C.Y, Ko, T.P, Wang, H.C, Chang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and functional analyses of viral H2 protein of the vaccinia virus entry fusion complex.
J.Virol., 97, 2023
|
|
2FEJ
 
 | | Solution structure of human p53 DNA binding domain. | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Perez-Canadillas, J.M, Tidow, H, Freund, S.M, Rutherford, T.J, Ang, H.C, Fersht, A.R. | | Deposit date: | 2005-12-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of p53 core domain: Structural basis for its instability
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6LYJ
 
 | | The crystal structure of SAUGI/EBVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-14 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
6LYV
 
 | | The crystal structure of SAUGI/KSHVUDG complex | | Descriptor: | SAUGI, Uracil-DNA glycosylase | | Authors: | Liao, Y.T, Ko, T.P, Wang, H.C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insight into the differential interactions between the DNA mimic protein SAUGI and two gamma herpesvirus uracil-DNA glycosylases.
Int.J.Biol.Macromol., 160, 2020
|
|
5XVE
 
 | |
6B8C
 
 | |
