1F90
 
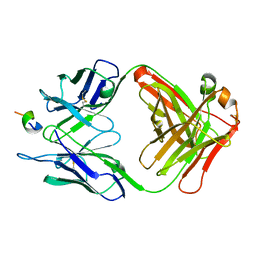 | | FAB FRAGMENT OF MONOCLONAL ANTIBODY (LNKB-2) AGAINST HUMAN INTERLEUKIN-2 IN COMPLEX WITH ANTIGENIC PEPTIDE | | Descriptor: | ANTIGENIC NONAPEPTIDE, FAB FRAGMENT OF MONOCLONAL ANTIBODY | | Authors: | Afonin, P.V, Fokin, A.V, Tsigannik, I.N, Mikhailova, I.Y, Onoprienko, L.V, Mikhaleva, I.I, Ivanov, V.T, Mareeva, T.Y, Nesmeyanov, V.A, Li, N, Duax, W.L, Pletnev, V.Z. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an anti-interleukin-2 monoclonal antibody Fab complexed with an antigenic nonapeptide.
Protein Sci., 10, 2001
|
|
5TIS
 
 | | Room temperature XFEL structure of the native, doubly-illuminated photosystem II complex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Fuller, F, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Hussein, R, Zhang, M, Douthit, L, Kubin, M, de Lichtenberg, C, Pham, L.V, Nilsson, H, Cheah, M.H, Shevela, D, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Pastor, E, Weninger, C, Fransson, T, Lassalle, L, Braeuer, P, Aller, P, Docker, P.T, Andi, B, Orville, A.M, Glownia, J.M, Nelson, S, Sikorski, M, Zhu, D, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Watermann, D.G, Evans, G, Wernet, P, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-10-03 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25000381 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5OF4
 
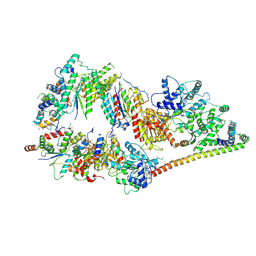 | | The cryo-EM structure of human TFIIH | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, General transcription factor IIH subunit 4,p52,General transcription factor IIH subunit 4, ... | | Authors: | Greber, B.J, Nguyen, T.H.D, Fang, J, Afonine, P.V, Adams, P.D, Nogales, E. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | The cryo-electron microscopy structure of human transcription factor IIH.
Nature, 549, 2017
|
|
3J40
 
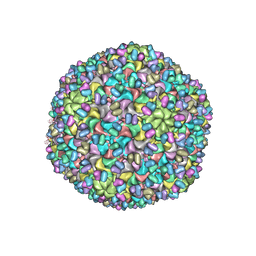 | | Validated Near-Atomic Resolution Structure of Bacteriophage Epsilon15 Derived from Cryo-EM and Modeling | | Descriptor: | gp10, gp7 | | Authors: | Baker, M.L, Hryc, C.F, Zhang, Q, Wu, W, Jakana, J, Haase-Pettingell, C, Afonine, P.V, Adams, P.D, King, J.A, Jiang, W, Chiu, W. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Validated near-atomic resolution structure of bacteriophage epsilon15 derived from cryo-EM and modeling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3J7L
 
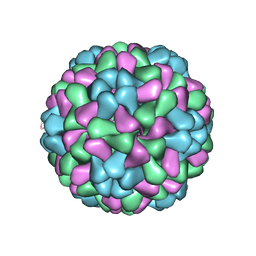 | | Full virus map of brome mosaic virus | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7N
 
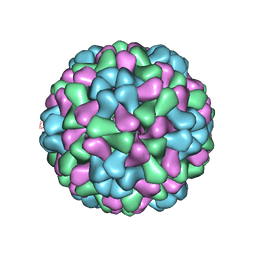 | | Virus model of brome mosaic virus (second half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3J7M
 
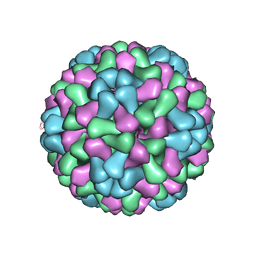 | | Virus model of brome mosaic virus (first half data set) | | Descriptor: | Capsid protein | | Authors: | Wang, Z, Hryc, C, Bammes, B, Afonine, P.V, Jakana, J, Chen, D.H, Liu, X, Baker, M.L, Kao, C, Ludtke, S.J, Schmid, M.F, Adams, P.D, Chiu, W. | | Deposit date: | 2014-07-18 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | An atomic model of brome mosaic virus using direct electron detection and real-space optimization.
Nat Commun, 5, 2014
|
|
3OS8
 
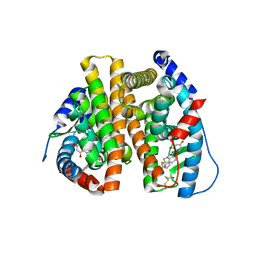 | | Estrogen Receptor | | Descriptor: | 4-[1-benzyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.031 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
3OSA
 
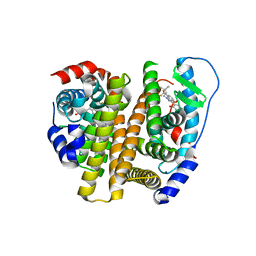 | | Estrogen Receptor | | Descriptor: | 4-[1-(3-methylbut-2-en-1-yl)-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.296 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
3OS9
 
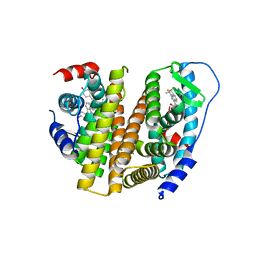 | | Estrogen Receptor | | Descriptor: | 4-[1-allyl-7-(trifluoromethyl)-1H-indazol-3-yl]benzene-1,3-diol, Estrogen receptor | | Authors: | Bruning, J, Parent, A.A, Gil, G, Zhao, M, Nowak, J, Pace, M.C, Smith, C.L, Afonine, P.V, Adams, P.D, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Coupling of receptor conformation and ligand orientation determine graded activity.
Nat.Chem.Biol., 6, 2010
|
|
5UU5
 
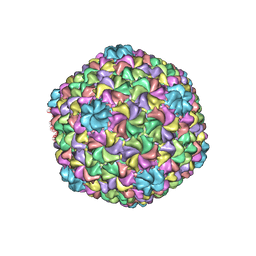 | | Bacteriophage P22 mature virion capsid protein | | Descriptor: | Major capsid protein | | Authors: | Hryc, C.F, Chen, D.-H, Afonine, P.V, Jakana, J, Wang, Z, Haase-Pettingell, C, Jiang, W, Adams, P.D, King, J.A, Schmid, M.F, Chiu, W. | | Deposit date: | 2017-02-16 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Accurate model annotation of a near-atomic resolution cryo-EM map.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WNW
 
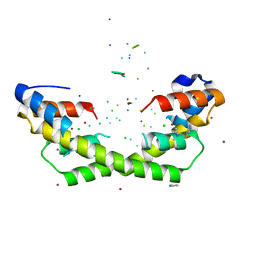 | | Chaperone Spy bound to Im7 6-45 ensemble | | Descriptor: | CHLORIDE ION, Colicin-E7 immunity protein, IMIDAZOLE, ... | | Authors: | Horowitz, S, Salmon, L, Koldewey, P, Ahlstrom, L.S, Martin, R, Xu, Q, Afonine, P.V, Trievel, R.C, Brooks, C.L, Bardwell, J.C.A. | | Deposit date: | 2017-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Visualizing chaperone-assisted protein folding.
Nat. Struct. Mol. Biol., 23, 2016
|
|
6DZ1
 
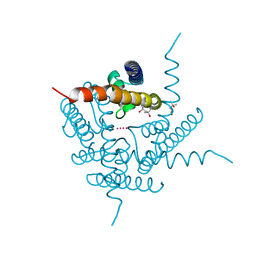 | | Studies of Ion Transport in K+ Channels | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein | | Authors: | Langan, P.S, Vandavasi, V.G, Weiss, K.L, Wagner, A, Duman, R, El Omari, K, Afonine, P.V, Coates, L. | | Deposit date: | 2018-07-02 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Anomalous X-ray diffraction studies of ion transport in K+channels.
Nat Commun, 9, 2018
|
|
1F8T
 
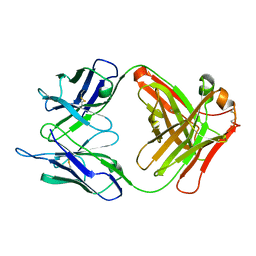 | | FAB (LNKB-2) OF MONOCLONAL ANTIBODY, CRYSTAL STRUCTURE | | Descriptor: | ANTIBODY FAB FRAGMENT (HEAVY CHAIN), ANTIBODY FAB FRAGMENT (LIGHT CHAIN) | | Authors: | Fokin, A.V, Afonin, P.V, Mikhailova, I.I, Tsygannik, I.N, Mareeva, T.I. | | Deposit date: | 2000-07-05 | | Release date: | 2001-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spatial structure of a Fab-fragment of a monoclonal antibody to human interleukin-2 in two crystalline forms at a resolution of 2.2 and 2.9 angstroms
Bioorg.Khim., 26, 2000
|
|
6EYC
 
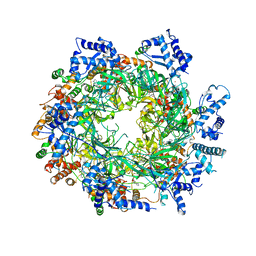 | | Re-refinement of the MCM2-7 double hexamer using ISOLDE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Croll, T.I. | | Deposit date: | 2017-11-11 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
4YE7
 
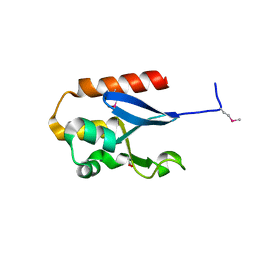 | |
5H31
 
 | |
4TNH
 
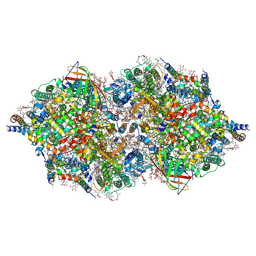 | | RT XFEL structure of Photosystem II in the dark state at 4.9 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.900007 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4TNL
 
 | | 1.8 A resolution room temperature structure of Thermolysin recorded using an XFEL | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
6VBK
 
 | | Crystal structure of N-terminal domain of Mycobacterium tuberculosis complex Lon protease | | Descriptor: | GLYCEROL, Lon211 | | Authors: | Bi, F.K, Chen, C, Chen, X.Y, Guo, C.Y, Lin, D.H. | | Deposit date: | 2019-12-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the N domain of Lon protease from Mycobacterium avium complex.
Protein Sci., 28, 2019
|
|
6UP0
 
 | |
6UTO
 
 | | Native E. coli Glyceraldehyde 3-phosphate dehydrogenase | | Descriptor: | ACETATE ION, Glyceraldehyde-3-phosphate dehydrogenase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Rodriguez-Hernandez, A, Romo-Arevalo, E, Rodriguez-Romero, A. | | Deposit date: | 2019-10-29 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A Novel Substrate-Binding Site in the X-Ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Crystals, 9, 2019
|
|
4TNJ
 
 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
