4HQX
 
 | | CRYSTAL STRUCTURE OF HUMAN PDGF-BB IN COMPLEX WITH A Modified nucleotide aptamer (SOMAmer SL4) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GO3
 
 | |
4GO4
 
 | | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative gamma-hydroxymuconic semialdehyde dehydrogenase | | Authors: | Su, J, Zhang, C, Liu, S, Zhu, D, Gu, L. | | Deposit date: | 2012-08-18 | | Release date: | 2013-09-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Crystal structure of PnpE in complex with Nicotinamide adenine dinucleotide
To be Published
|
|
4HQU
 
 | | Crystal structure of human PDGF-BB in complex with a modified nucleotide aptamer (SOMAmer SL5) | | Descriptor: | MAGNESIUM ION, Platelet-derived growth factor subunit B, SODIUM ION, ... | | Authors: | Davies, D.R, Edwards, T.E, Janjic, N, Gelinas, A.D, Zhang, C, Jarvis, T.C. | | Deposit date: | 2012-10-26 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4LA4
 
 | |
4LAF
 
 | |
6C1R
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MALONATE ION, OLEIC ACID, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D26
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
3D9H
 
 | | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein | | Descriptor: | cDNA FLJ77766, highly similar to Homo sapiens ankyrin repeat and SOCS box-containing 9 (ASB9), transcript variant 2, ... | | Authors: | Fei, X, Gu, X, Fan, S, Zhang, C, Ji, C. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Splice Variant of Human ASB9 (hASB9-2), an Ankyrin Repeat Protein
To be published
|
|
6NSQ
 
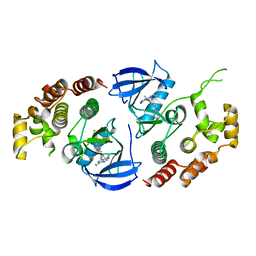 | | Crystal structure of BRAF kinase domain bound to the inhibitor 2l | | Descriptor: | 5-[(4-amino-1-ethyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)ethynyl]-N-(4-chlorophenyl)-6-methylisoquinolin-1-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Zhang, C, Sicheri, F. | | Deposit date: | 2019-01-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rigidification Dramatically Improves Inhibitor Selectivity for RAF Kinases.
Acs Med.Chem.Lett., 10, 2019
|
|
8G4G
 
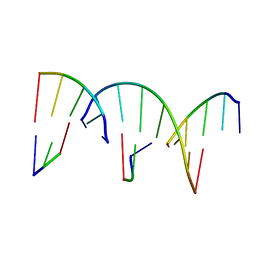 | | Crystal Engineering with One 8-mer DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*GP*G)-3'), DNA (5'-D(*AP*TP*CP*GP*GP*CP*CP*G)-3'), DNA (5'-D(P*CP*CP*G)-3') | | Authors: | Zhao, J, Zhang, C, Lu, B, Seeman, N.C, Noinaj, N, Sha, R, Mao, C. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Divergence and Convergence: Complexity Emerges in Crystal Engineering from an 8-mer DNA.
J.Am.Chem.Soc., 145, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID6
 
 | | Cryo-EM structure of the oleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID4
 
 | | Cryo-EM structure of the linoleic acid bound GPR120-Gi complex | | Descriptor: | Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8CX6
 
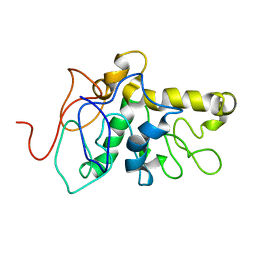 | | TPX2 Minimal Active Domain on Microtubules | | Descriptor: | Targeting protein for Xklp2-A | | Authors: | Guo, C, Alfaro-Aco, R, Russell, R, Zhang, C, Petry, S, Polenova, T. | | Deposit date: | 2022-05-19 | | Release date: | 2023-06-28 | | Method: | SOLID-STATE NMR | | Cite: | Structural basis of protein condensation on microtubules underlying branching microtubule nucleation.
Nat Commun, 14, 2023
|
|
5Z3Q
 
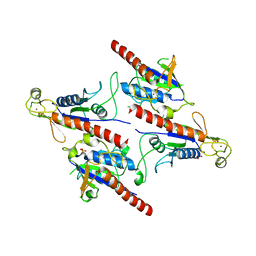 | | Crystal Structure of a Soluble Fragment of Poliovirus 2C ATPase (2.55 Angstrom) | | Descriptor: | PHOSPHATE ION, PV-2C, ZINC ION | | Authors: | Guan, H, Tian, J, Zhang, C, Qin, B, Cui, S. | | Deposit date: | 2018-01-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Crystal structure of a soluble fragment of poliovirus 2CATPase
PLoS Pathog., 14, 2018
|
|
5L6P
 
 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 6) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ephrin type-B receptor 3, ~{N}-(4-phenylazanylquinazolin-7-yl)ethanamide | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
7Y36
 
 | | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, HYDROXIDE ION, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-10 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the Teriparatide-bound human PTH1R-Gs complex
to be published
|
|
7Y35
 
 | | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex | | Descriptor: | Abaloparatide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhai, X, Mao, C, Shen, Q, Zang, S, Shen, D, Zhang, H, Chen, Z, Wang, G, Zhang, C, Zhang, Y, Liu, Z. | | Deposit date: | 2022-06-09 | | Release date: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the Abaloparatide-bound human PTH1R-Gs complex
To Be Published
|
|
5L6O
 
 | | EphB3 kinase domain covalently bound to an irreversible inhibitor (compound 3) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 1-(4-phenylazanylquinazolin-7-yl)ethanone, Ephrin type-B receptor 3 | | Authors: | Schimpl, M, Overman, R, Kung, A, Chen, Y.-C, Ni, F, Zhu, J, Turner, M, Molina, H, Zhang, C. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Development of Specific, Irreversible Inhibitors for a Receptor Tyrosine Kinase EphB3.
J.Am.Chem.Soc., 138, 2016
|
|
5MJB
 
 | | Kinase domain of human EphB1, G703C mutant, covalently bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
5MJA
 
 | | Kinase domain of human EphB1 bound to a quinazoline-based inhibitor | | Descriptor: | 2-chloranyl-~{N}-[4-[(2-chloranyl-5-oxidanyl-phenyl)amino]quinazolin-7-yl]ethanamide, Ephrin type-B receptor 1, SULFATE ION | | Authors: | Kung, A, Schimpl, M, Chen, Y.-C, Overman, R.C, Zhang, C. | | Deposit date: | 2016-11-30 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A Chemical-Genetic Approach to Generate Selective Covalent Inhibitors of Protein Kinases.
ACS Chem. Biol., 12, 2017
|
|
