7WP8
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK1628x in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2B4 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | Deposit date: | 2022-01-23 | | Release date: | 2023-03-01 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
4LYN
 
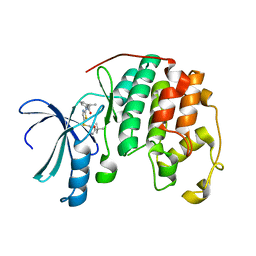 | | Crystal structure of cyclin-dependent kinase 2 (cdk2-wt) complex with (2s)-n-(5-(((5-tert-butyl-1,3-oxazol-2-yl)methyl)sulfanyl)-1,3-thiazol-2-yl)-2-phenylpropanamide | | Descriptor: | (2S)-N-(5-{[(5-tert-butyl-1,3-oxazol-2-yl)methyl]sulfanyl}-1,3-thiazol-2-yl)-2-phenylpropanamide, Cyclin-dependent kinase 2 | | Authors: | Sack, J.S. | | Deposit date: | 2013-07-31 | | Release date: | 2013-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Aminothiazole Inhibitors of Cyclin-Dependent Kinase 2: Synthesis, X-Ray Crystallographic Analysis, and Biological Activities
J.Med.Chem., 45, 2002
|
|
4CIN
 
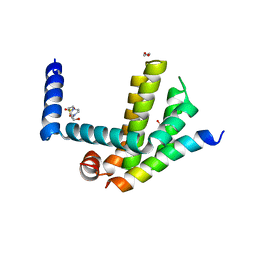 | | Complex of Bcl-xL with its BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
4CIM
 
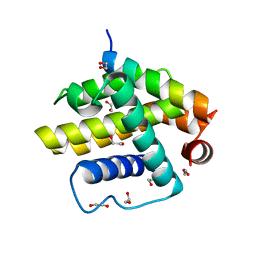 | | Complex of a Bcl-w BH3 mutant with a BH3 domain | | Descriptor: | 1,2-ETHANEDIOL, BCL-2-LIKE PROTEIN 2 | | Authors: | Colman, P.M, Lee, E.F, Fairlie, W.D. | | Deposit date: | 2013-12-12 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Functional Differences of Pro-Survival and Pro-Apoptotic B Cell Lymphoma 2 (Bcl-2) Proteins Depend on Structural Differences in Their Bcl-2 Homology 3 (Bh3) Domains
J.Biol.Chem., 289, 2014
|
|
4HVL
 
 | |
2QZK
 
 | | Crystal structure of human Beta Secretase complexed with I21 | | Descriptor: | 2-[(5R)-5-amino-5-methyl-4,16-dioxo-14-phenyl-3-oxa-15-azatricyclo[15.3.1.1~7,11~]docosa-1(21),7(22),8,10,12,14,17,19-octaen-19-yl]benzonitrile, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2007-08-16 | | Release date: | 2008-04-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategies toward improving the brain penetration of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
6KDY
 
 | |
6BNT
 
 | |
6ASY
 
 | | BiP-ATP2 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Liu, Q, Yang, J, Zong, Y, Columbus, L, Zhou, L. | | Deposit date: | 2017-08-26 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.
Nat Commun, 8, 2017
|
|
8G5H
 
 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8FOI
 
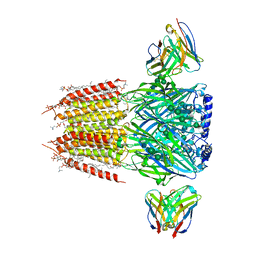 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2022-12-30 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4O
 
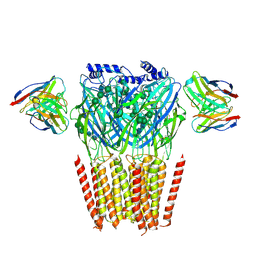 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with didesethylflurazepam and endogenous GABA | | Descriptor: | (5M)-1-(2-aminoethyl)-7-chloro-5-(2-fluorophenyl)-1,3-dihydro-2H-1,4-benzodiazepin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4N
 
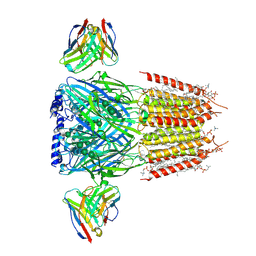 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroids | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, DODECANE, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5F
 
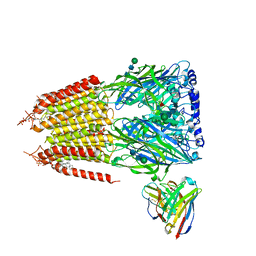 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5G
 
 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4X
 
 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
5TEG
 
 | |
6Y06
 
 | |
6Y07
 
 | |
5W3D
 
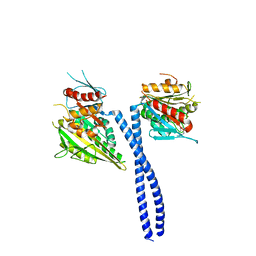 | | The structure of kinesin-14 wild-type Ncd-ADP dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Protein claret segregational | | Authors: | Park, H.W, Ma, Z, Chacko, J, Jiang, S.M, Robinson, R.C, Endow, S.A. | | Deposit date: | 2017-06-07 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
5W8J
 
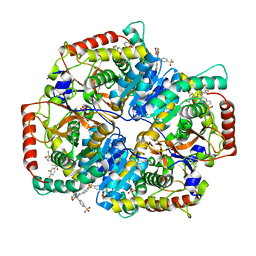 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 | | Descriptor: | 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Lukacs, C.M, Moulin, A. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8K
 
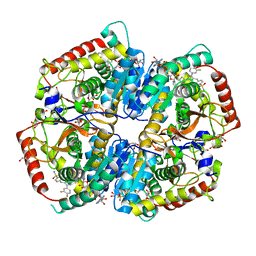 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 29 and NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-{3-(3,4-difluorophenyl)-5-hydroxy-4-[(4-sulfamoylphenyl)methyl]-1H-pyrazol-1-yl}-1,3-thiazole-4-carboxylic acid, GLYCEROL, ... | | Authors: | Lukacs, C.M, Dranow, D.M. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
5W8I
 
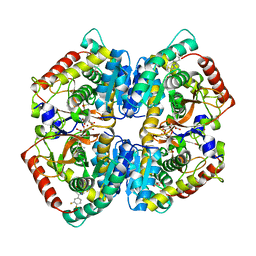 | | Crystal Structure of Lactate Dehydrogenase A in complex with inhibitor compound 23 and Zinc | | Descriptor: | 2-[3-(3,4-difluorophenyl)-5-hydroxy-1H-pyrazol-1-yl]-1,3-thiazole-4-carboxylic acid, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Lukacs, C.M, Abendroth, J. | | Deposit date: | 2017-06-21 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of Potent, Cell-Active Pyrazole-Based Inhibitors of Lactate Dehydrogenase (LDH).
J. Med. Chem., 60, 2017
|
|
7TO4
 
 | | Structural and functional impact by SARS-CoV-2 Omicron spike mutations | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiao, T.S, Cai, Y.F, Peng, H.Q, Volloch, S.R, Chen, B. | | Deposit date: | 2022-01-22 | | Release date: | 2022-02-16 | | Last modified: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural and functional impact by SARS-CoV-2 Omicron spike mutations.
Cell Rep, 39, 2022
|
|
