7OLS
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl pyrazole group | | Descriptor: | 5-[4-(1,5-dimethylpyrazol-4-yl)-2-methyl-phenyl]-~{N}-(imidazo[1,2-a]pyridin-6-ylmethyl)-~{N}-methyl-1,3,4-oxadiazol-2-amine, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLV
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a di-methyl, cyano pyrazole group | | Descriptor: | 4-[4-[5-[imidazo[1,2-a]pyridin-6-ylmethyl(methyl)amino]-1,3,4-oxadiazol-2-yl]-3-methyl-phenyl]-2,5-dimethyl-pyrazole-3-carbonitrile, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
7OLX
 
 | | MerTK kinase domain with type 1.5 inhibitor containing a tri-methyl pyrazole group | | Descriptor: | CHLORIDE ION, Tyrosine-protein kinase Mer, ~{N}-[[3-[4-[(dimethylamino)methyl]phenyl]imidazo[1,2-a]pyridin-6-yl]methyl]-~{N}-methyl-5-[3-methyl-5-(1,3,5-trimethylpyrazol-4-yl)pyridin-2-yl]-1,3,4-oxadiazol-2-amine | | Authors: | Pflug, A, Schimpl, M, McCoull, W, Nissink, J.W.M, Winter-Holt, J. | | Deposit date: | 2021-05-20 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Optimization of an Imidazo[1,2- a ]pyridine Series to Afford Highly Selective Type I1/2 Dual Mer/Axl Kinase Inhibitors with In Vivo Efficacy.
J.Med.Chem., 64, 2021
|
|
8WE9
 
 | | Human L-type voltage-gated calcium channel Cav1.2 (Class I) in the presence of pinaverium at 3.0 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE7
 
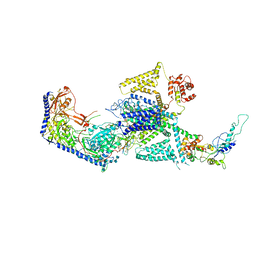 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WEA
 
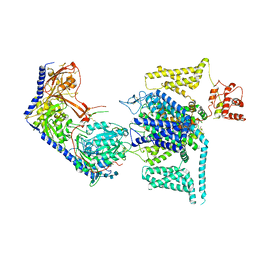 | | Human L-type voltage-gated calcium channel Cav1.2 (Class II) in the presence of pinaverium at 3.2 Angstrom resolution | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Fan, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE6
 
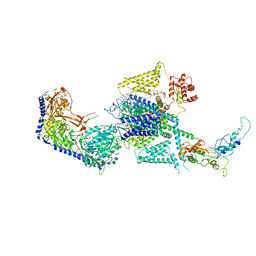 | | Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8WE8
 
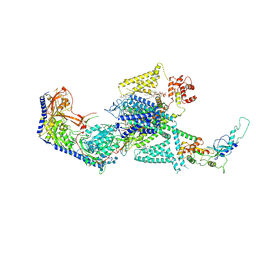 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine, amlodipine and pinaverium at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
8W4U
 
 | | human KCNQ2-CaM in complex with PIP2 and HN37 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
5I78
 
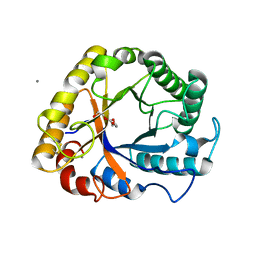 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
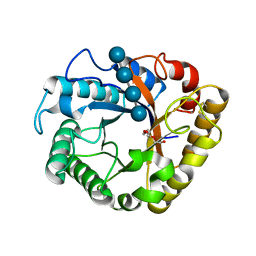 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I77
 
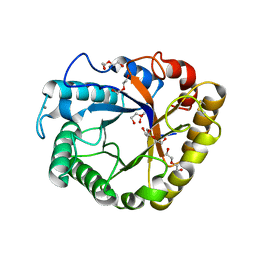 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
6D06
 
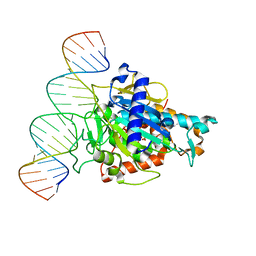 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
4R28
 
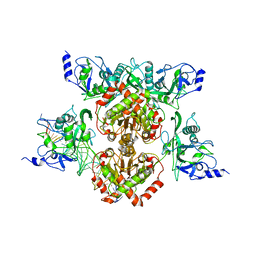 | |
4XZ7
 
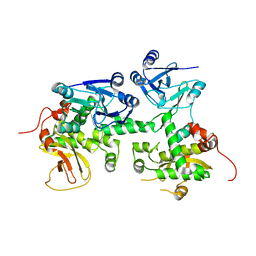 | | Crystal structure of a TGase | | Descriptor: | Putative uncharacterized protein | | Authors: | Yu, J, Ge, J, Yang, M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis
J.Biol.Chem., 290, 2015
|
|
8DPX
 
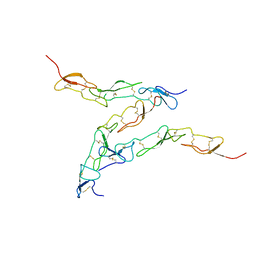 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
3HC8
 
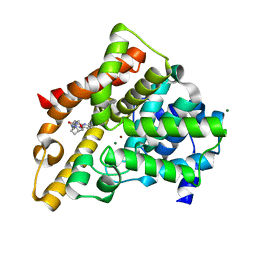 | | Investigation of Aminopyridiopyrazinones as PDE5 Inhibitors: Evaluation of Modifications to the Central Ring System. | | Descriptor: | 6-(6-methoxypyridin-3-yl)-2-[(2-morpholin-4-ylethyl)amino]-4-(2-propoxyethyl)pyrido[2,3-b]pyrazin-3(4H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Hughes, R.O, Stallings, W.C, Cubbage, J.W, Williams, J.M. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-07 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Investigation of aminopyridiopyrazinones as PDE5 inhibitors: Evaluation of modifications to the central ring system.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
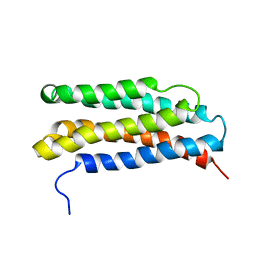
1LS4
 
 | |
5KL4
 
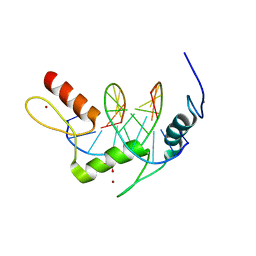 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with formylated DNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL6
 
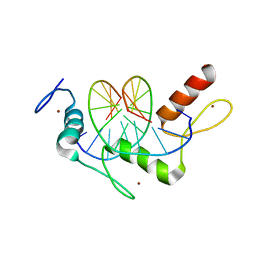 | | Wilms Tumor Protein (WT1) Q369R ZnF2-4 in complex with DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL5
 
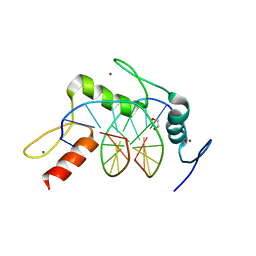 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL2
 
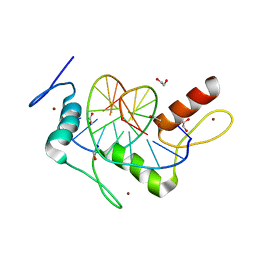 | | Wilms Tumor Protein (WT1) ZnF2-4 in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL3
 
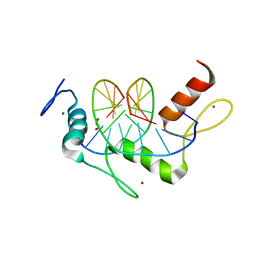 | | Wilms Tumor Protein (WT1) ZnF2-4 Q369H in complex with DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*AP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
5KL7
 
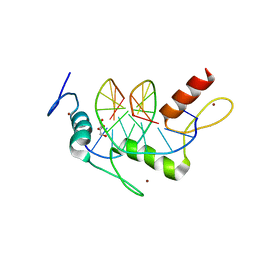 | | Wilms Tumor Protein (WT1) ZnF2-4Q369R in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Cheng, X. | | Deposit date: | 2016-06-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.579 Å) | | Cite: | Denys-Drash syndrome associated WT1 glutamine 369 mutants have altered sequence-preferences and altered responses to epigenetic modifications.
Nucleic Acids Res., 44, 2016
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2014-04-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
