4J00
 
 | | Crystal Structure of Fischerella Transcription Factor HetR complexed with 24mer DNA target | | Descriptor: | DNA (5'-D(*TP*GP*GP*TP*GP*AP*GP*GP*GP*GP*TP*TP*AP*AP*AP*CP*CP*CP*CP*TP*CP*AP*CP*C)-3'), MAGNESIUM ION, Transcription Factor HetR | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7YE9
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDI
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2, focused refinement of RBD region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32, Light chain of R1-32, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YDY
 
 | | SARS-CoV-2 Spike (6P) in complex with 1 R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
7YEG
 
 | | SARS-CoV-2 Spike (6P) in complex with 3 R1-32 Fabs and 3 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, X, He, J, Chen, L, Xiong, X. | | Deposit date: | 2022-07-05 | | Release date: | 2022-08-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | SARS-CoV-2 Delta and Omicron variants evade population antibody response by mutations in a single spike epitope.
Nat Microbiol, 7, 2022
|
|
5WMT
 
 | |
4ONM
 
 | | Crystal structure of human Mms2/Ubc13 - NSC697923 | | Descriptor: | 2-[(4-methylphenyl)sulfonyl]-5-nitrofuran, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUM
 
 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CITRATE ANION, Caprin-2, ISOPROPYL ALCOHOL, ... | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4ONN
 
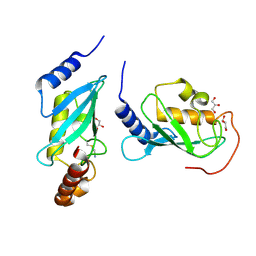 | | Crystal structure of human Mms2/Ubc13 - BAY 11-7082 | | Descriptor: | 3-[(4-methylphenyl)sulfonyl]prop-2-enenitrile, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUS
 
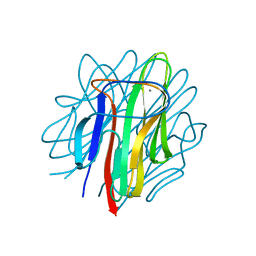 | | Crystal structure of zebrafish Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2 | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-18 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
4ONL
 
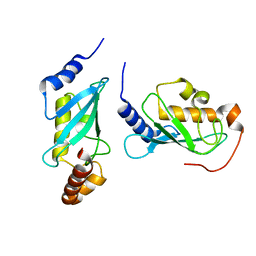 | | Crystal structure of human Mms2/Ubc13_D81N, R85S, A122V, N123P | | Descriptor: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUL
 
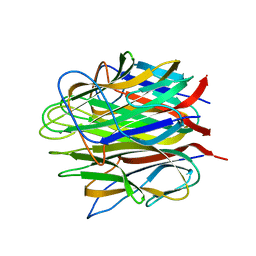 | | Crystal structure of human Caprin-2 C1q domain | | Descriptor: | CALCIUM ION, Caprin-2, GLYCEROL | | Authors: | Song, X, Li, L. | | Deposit date: | 2014-02-17 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling
J.Biol.Chem., 289, 2014
|
|
6BAW
 
 | | Structure of GRP94 with a selective resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmin, PHOSPHATE ION, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
6C91
 
 | | Structure of GRP94 with a resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, 5-[2-(1-benzyl-1H-imidazol-2-yl)ethyl]-4,6-dichlorobenzene-1,3-diol, Endoplasmin, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2018-01-25 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.895 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
7CL1
 
 | | Human SIRT6 in complex with allosteric activator MDL-801 (3.2A) | | Descriptor: | 5-[[3,5-bis(chloranyl)phenyl]sulfonylamino]-2-[(5-bromanyl-4-fluoranyl-2-methyl-phenyl)sulfamoyl]benzoic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2021-05-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
2JG4
 
 | | Substrate-free IDE structure in its closed conformation | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
2JBU
 
 | | Crystal structure of human insulin degrading enzyme complexed with co- purified peptides. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, CO-PURIFIED PEPTIDE, INSULIN-DEGRADING ENZYME | | Authors: | Im, H, Shen, Y, Tang, W.J. | | Deposit date: | 2006-12-11 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
7VAH
 
 | |
7EMN
 
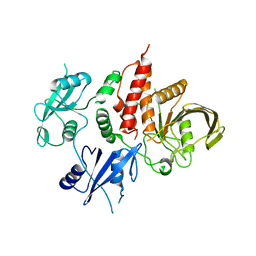 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
8IVQ
 
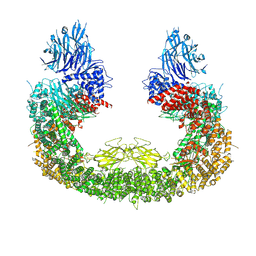 | | Cryo-EM structure of mouse BIRC6, Global map | | Descriptor: | Isoform 2 of Baculoviral IAP repeat-containing protein 6 | | Authors: | Liu, S, Jiang, T, Bu, F, Zhao, J, Wang, G, Li, N, Gao, N, Qiu, X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanisms underlying the BIRC6-mediated regulation of apoptosis and autophagy.
Nat Commun, 15, 2024
|
|
2MDJ
 
 | |
2MDI
 
 | | Solution structure of the PP2WW mutant (KPP2WW) of HYPB | | Descriptor: | Histone-lysine N-methyltransferase SETD2 | | Authors: | Gao, Y.G, Hu, H.Y. | | Deposit date: | 2013-09-10 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of the WW domain of HYPB/SETD2 regulates its interaction with the proline-rich region of huntingtin.
Structure, 22, 2014
|
|
