4TTP
 
 | |
4TTQ
 
 | |
7JR9
 
 | | Chlamydomonas reinhardtii radial spoke minimal head complex | | Descriptor: | Flagellar radial spoke protein 10, Flagellar radial spoke protein 4, Flagellar radial spoke protein 6, ... | | Authors: | Grossman-Haham, I, Coudray, N, Yu, Z, Wang, F, Bhabha, G, Vale, R.D. | | Deposit date: | 2020-08-11 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the radial spoke head and insights into its role in mechanoregulation of ciliary beating.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7CYV
 
 | | Crystal structure of FD20, a neutralizing single-chain variable fragment (scFv) in complex with SARS-CoV-2 Spike receptor-binding domain (RBD) | | Descriptor: | Spike protein S1, The heavy chain variable region of the scFv FD20,The light chain variable region of the scFv FD20, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)][alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, Y, Li, T, Lai, Y, Cai, H, Yao, H, Li, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Uncovering a conserved vulnerability site in SARS-CoV-2 by a human antibody.
Embo Mol Med, 13, 2021
|
|
7E5Y
 
 | | Molecular basis for neutralizing antibody 2B11 targeting SARS-CoV-2 RBD | | Descriptor: | 2B11 Fab Heavy chain, 2B11 Fab Light chain, Spike protein S1 | | Authors: | Wu, H, Yu, F, Wang, Q.S, Zhou, H, Wang, W.W, Zhao, T, Pan, Y.B, Yang, X.M. | | Deposit date: | 2021-02-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Screening of potent neutralizing antibodies against SARS-CoV-2 using convalescent patients-derived phage-display libraries.
Cell Discov, 7, 2021
|
|
7EVS
 
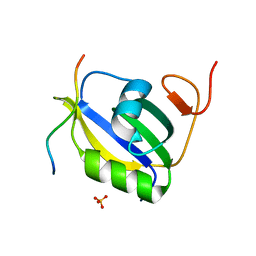 | | Crystal structure of hnRNP LL RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L-like, SHI domain from Histone-lysine N-methyltransferase SETD2, SULFATE ION | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7EVR
 
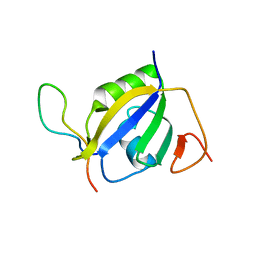 | | Crystal structure of hnRNP L RRM2 in complex with SETD2 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein L, SHI domain from Histone-lysine N-methyltransferase SETD2 | | Authors: | Li, F.D, Wang, S.M. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of the interaction between SETD2 methyltransferase and hnRNP L paralogs for governing co-transcriptional splicing.
Nat Commun, 12, 2021
|
|
7KBB
 
 | |
7KBA
 
 | |
7YJI
 
 | |
7XXL
 
 | | RBD in complex with Fab14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab14 heavy chain, Fab14 light chain, ... | | Authors: | Lin, J.Q, Tan, Y.J.E, Wu, B, Lescar, J. | | Deposit date: | 2022-05-30 | | Release date: | 2022-09-14 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Engineering SARS-CoV-2 specific cocktail antibodies into a bispecific format improves neutralizing potency and breadth.
Nat Commun, 13, 2022
|
|
7YR0
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (interface) | | Descriptor: | Heavy chain of S309, IGK@ protein, Spike protein S1, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQU
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQT
 
 | | SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQV
 
 | | pH 5.5 SARS-CoV-2 BA.2.75 S Trimer (1 RBD Up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR1
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with XG2v024 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQX
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR3
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YR2
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with ACE2(state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQW
 
 | | SARS-CoV-2 BA.2.75 S Trimer (3 RBD Down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQY
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YQZ
 
 | | SARS-CoV-2 BA.2.75 S Trimer in complex with S309 (state3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S309 heavy chain, S309 light chain, ... | | Authors: | Wang, L. | | Deposit date: | 2022-08-08 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75.
Cell Host Microbe, 30, 2022
|
|
7YF5
 
 | |
7CIR
 
 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
