5U77
 
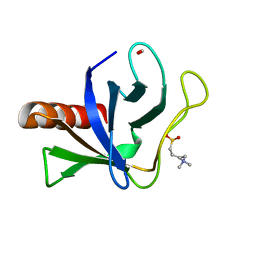 | | Crystal structure of ORP8 PH domain | | Descriptor: | FORMIC ACID, N-(2-hydroxyethyl)-N,N-dimethyl-3-sulfopropan-1-aminium, Oxysterol-binding protein-related protein 8 | | Authors: | Ghai, R, Yang, H. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.157 Å) | | Cite: | ORP5 and ORP8 bind phosphatidylinositol-4, 5-biphosphate (PtdIns(4,5)P 2) and regulate its level at the plasma membrane.
Nat Commun, 8, 2017
|
|
5C56
 
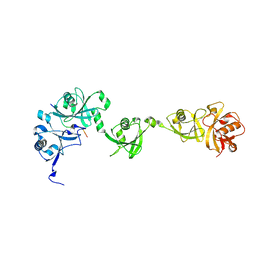 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
2VTY
 
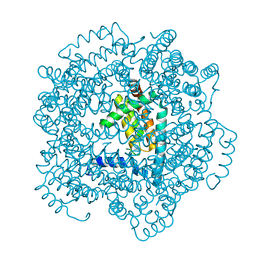 | | Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain swapped dimer | | Descriptor: | PROTEIN F1 | | Authors: | Kvansakul, M, Yang, H, Fairlie, W.D, Czabotar, P.E, Fischer, S.F, Perugini, M.A, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2008-05-19 | | Release date: | 2008-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vaccinia Virus Anti-Apoptotic F1L is a Novel Bcl-2-Like Domain-Swapped Dimer that Binds a Highly Selective Subset of Bh3-Containing Death Ligands.
Cell Death Differ., 15, 2008
|
|
5K83
 
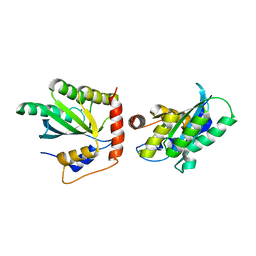 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K81
 
 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5K82
 
 | | Crystal Structure of a Primate APOBEC3G N-Terminal Domain | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.913 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
5KK5
 
 | | AsCpf1(E993A)-crRNA-DNA ternary complex | | Descriptor: | CRISPR-associated endonuclease Cpf1, DNA (28-MER), DNA (8-mer), ... | | Authors: | Gao, P, Yang, H, Rajashankar, K.R, Huang, Z, Patel, D.J. | | Deposit date: | 2016-06-21 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.289 Å) | | Cite: | Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition.
Cell Res., 26, 2016
|
|
3IU3
 
 | | Crystal structure of the Fab fragment of therapeutic antibody Basiliximab in complex with IL-2Ra (CD25) ectodomain | | Descriptor: | Heavy chain of Fab fragment of Basiliximab, Interleukin-2 receptor alpha chain, Light chain of Fab fragment of Basiliximab, ... | | Authors: | Du, J, Yang, H, Wang, J, Ding, J. | | Deposit date: | 2009-08-29 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the blockage of IL-2 signaling by therapeutic antibody basiliximab
J.Immunol., 184, 2010
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
8H3L
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (T21I and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3K
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Double Mutant (L50F and E166V) in Complex with Inhibitor Enstrelvir | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, ... | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8H3G
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166V Mutant in Complex with Inhibitor Enstrelvir | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione, GLYCEROL | | Authors: | Wang, H, Lin, M, Duan, Y, Zhang, X, Zhou, H, Bian, Q, Liu, X, Rao, Z, Yang, H. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanisms of SARS-CoV-2 resistance to nirmatrelvir.
Nature, 622, 2023
|
|
8HEC
 
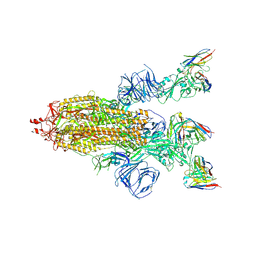 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
8HEB
 
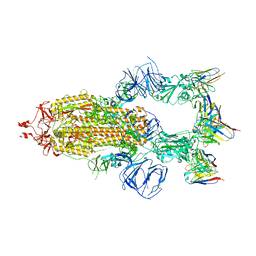 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
4PY0
 
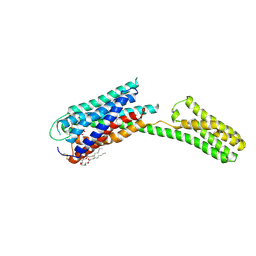 | | Crystal structure of P2Y12 receptor in complex with 2MeSATP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(tetrahydrogen triphosphate), P2Y purinoceptor 12, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4PXZ
 
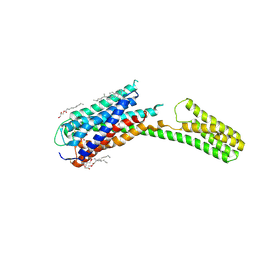 | | Crystal structure of P2Y12 receptor in complex with 2MeSADP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(methylsulfanyl)adenosine 5'-(trihydrogen diphosphate), CHOLESTEROL, ... | | Authors: | Zhang, J, Zhang, K, Gao, Z.G, Paoletta, S, Zhang, D, Han, G.W, Li, T, Ma, L, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Agonist-bound structure of the human P2Y12 receptor
Nature, 509, 2014
|
|
4QNP
 
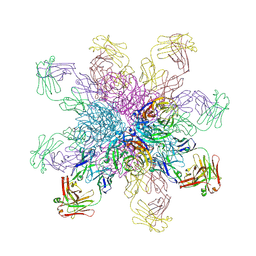 | | Crystal structure of the 2009 pandemic H1N1 influenza virus neuraminidase with a neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wan, H.Q, Yang, H, Shore, D.A, Garten, R.J, Couzens, L, Gao, J, Jiang, L.L, Carney, P.J, Villanueva, J, Stevens, J, Eichelberger, M.C. | | Deposit date: | 2014-06-18 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural characterization of a protective epitope spanning A(H1N1)pdm09 influenza virus neuraminidase monomers.
Nat Commun, 6, 2015
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
8PUA
 
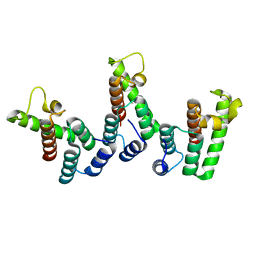 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUF
 
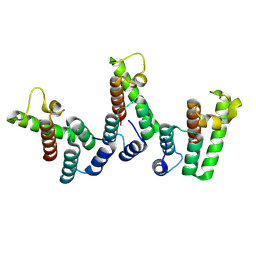 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUB
 
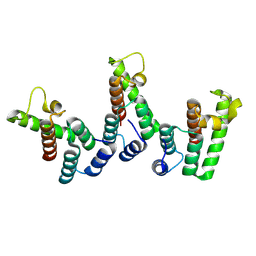 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUC
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUD
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
8PUG
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-NTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Unconventional stabilization of the human T-cell leukemia virus type 1 immature Gag lattice.
Biorxiv, 2023
|
|
