3K1E
 
 | | Crystal structure of odorant binding protein 1 (AaegOBP1) from Aedes aegypti | | 分子名称: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Leite, N.R, Krogh, R, Leal, W.S, Iulek, J, Oliva, G. | | 登録日 | 2009-09-27 | | 公開日 | 2009-12-08 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive "Lid".
Plos One, 4, 2009
|
|
3KZ5
 
 | | Structure of cdomain | | 分子名称: | ACETATE ION, Protein sopB | | 著者 | Schumacher, M.A. | | 登録日 | 2009-12-07 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3LQ8
 
 | | Structure of the kinase domain of c-Met bound to XL880 (GSK1363089) | | 分子名称: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | 著者 | Lougheed, J.C, Stout, T.J. | | 登録日 | 2010-02-08 | | 公開日 | 2010-05-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases.
Cancer Res., 69, 2009
|
|
3M9A
 
 | |
3MKZ
 
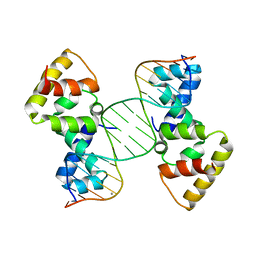 | | Structure of SopB(155-272)-18mer complex, P21 form | | 分子名称: | CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*GP*AP*CP*CP*AP*TP*GP*GP*TP*CP*CP*CP*AP*G)-3'), Protein sopB | | 著者 | Schumacher, M.A. | | 登録日 | 2010-04-15 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
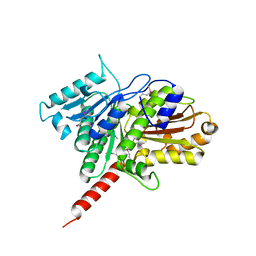
3M8K
 
 | |
3M8F
 
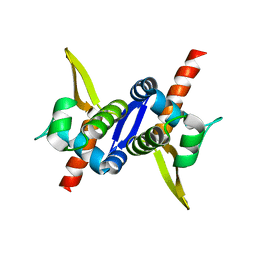 | |
3M8E
 
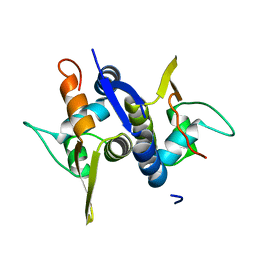 | |
3O52
 
 | | Structure of the E.coli GDP-mannose hydrolase (yffh) in complex with tartrate | | 分子名称: | CHLORIDE ION, D(-)-TARTARIC ACID, GDP-mannose pyrophosphatase nudK, ... | | 著者 | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | 登録日 | 2010-07-27 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O6Z
 
 | | Structure of the D152A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | 登録日 | 2010-07-29 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O69
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with Mg++ | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GDP-mannose pyrophosphatase nudK, ... | | 著者 | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | 登録日 | 2010-07-28 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
3O61
 
 | | Structure of the E100A E.coli GDP-mannose hydrolase (yffh) in complex with GDP-mannose and Mg++ | | 分子名称: | CHLORIDE ION, GDP-mannose pyrophosphatase nudK, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | 著者 | Amzel, L.M, Gabelli, S.B, Boto, A.N. | | 登録日 | 2010-07-28 | | 公開日 | 2011-05-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structural studies of the Nudix GDP-mannose hydrolase from E. coli reveals a new motif for mannose recognition.
Proteins, 79, 2011
|
|
8K3K
 
 | |
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | 分子名称: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-08-29 | | 公開日 | 2014-10-01 | | 最終更新日 | 2016-11-02 | | 実験手法 | X-RAY DIFFRACTION (1.659 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | 著者 | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.85 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | 著者 | Wang, K, Wang, X, Pan, L. | | 登録日 | 2021-08-07 | | 公開日 | 2021-10-20 | | 最終更新日 | 2022-07-06 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | 分子名称: | ACETATE ION, Oxidoreductase, SULFATE ION | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-09 | | 公開日 | 2012-11-28 | | 最終更新日 | 2016-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
8I4T
 
 | | Structure of the asymmetric unit of SFTSV virion | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | 著者 | Du, S, Peng, R, Qi, J, Li, C. | | 登録日 | 2023-01-21 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (5.2 Å) | | 主引用文献 | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
4XAU
 
 | | Crystal structure of AtS13 from Actinomadura melliaura | | 分子名称: | PYRIDOXAL-5'-PHOSPHATE, Putative aminotransferase | | 著者 | Wang, F, Singh, S, Xu, W, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-12-15 | | 公開日 | 2014-12-24 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.0012 Å) | | 主引用文献 | Structural characterization of AtmS13, a putative sugar aminotransferase involved in indolocarbazole AT2433 aminopentose biosynthesis.
Proteins, 83, 2015
|
|
8ILQ
 
 | | Structure of SFTSV Gn-Gc heterodimer | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | 著者 | Du, S, Peng, R, Qi, J, Li, C. | | 登録日 | 2023-03-04 | | 公開日 | 2023-09-13 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8JVE
 
 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | 分子名称: | 1,2-ETHANEDIOL, 1-(3-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | 著者 | Anantharajan, J, Baburajendran, N. | | 登録日 | 2023-06-28 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
8JVL
 
 | | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer | | 分子名称: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-1,2,3,4-tetrazole, Ubiquitin-conjugating enzyme E2 T | | 著者 | Anantharajan, J, Baburajendran, N. | | 登録日 | 2023-06-28 | | 公開日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Identification and characterization of inhibitors covalently modifying catalytic cysteine of UBE2T and blocking ubiquitin transfer.
Biochem.Biophys.Res.Commun., 689, 2023
|
|
1I7X
 
 | | BETA-CATENIN/E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, EPITHELIAL-CADHERIN | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I7W
 
 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
7E3L
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 58G6 heavy chain, 58G6 light chain, ... | | 著者 | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | 登録日 | 2021-02-09 | | 公開日 | 2021-09-15 | | 最終更新日 | 2022-03-02 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
