3HZI
 
 | | Structure of mdt protein | | 分子名称: | 5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3', ADENOSINE-5'-TRIPHOSPHATE, HTH-type transcriptional regulator hipB, ... | | 著者 | Schumacher, M.A. | | 登録日 | 2009-06-23 | | 公開日 | 2009-07-28 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.98 Å) | | 主引用文献 | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
5ZUV
 
 | | Crystal Structure of the Human Coronavirus 229E HR1 motif in complex with pan-CoVs inhibitor EK1 | | 分子名称: | CHLORIDE ION, Spike glycoprotein,Spike glycoprotein,inhibitor EK1 | | 著者 | Yan, L, Yang, B, Wilson, I.A. | | 登録日 | 2018-05-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike.
Sci Adv, 5, 2019
|
|
5ZVM
 
 | |
5ZVK
 
 | |
6B85
 
 | | Crystal structure of transmembrane protein TMHC4_R | | 分子名称: | TMHC4_R | | 著者 | Lu, P, DiMaio, F, Min, D, Bowie, J, Wei, K.Y, Baker, D. | | 登録日 | 2017-10-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (3.889 Å) | | 主引用文献 | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
6B87
 
 | | Crystal structure of transmembrane protein TMHC2_E | | 分子名称: | TMHC2_E | | 著者 | Lu, P, DiMaio, F, Min, D, Wei, K.Y, Bowie, J, Baker, D. | | 登録日 | 2017-10-05 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.947 Å) | | 主引用文献 | Accurate computational design of multipass transmembrane proteins.
Science, 359, 2018
|
|
4HFQ
 
 | | Crystal structure of UDP-X diphosphatase | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Duong-Ly, K.C, Amzel, L.M, Gabelli, S.B. | | 登録日 | 2012-10-05 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.39 Å) | | 主引用文献 | A UDP-X diphosphatase from Streptococcus pneumoniae hydrolyzes precursors of peptidoglycan biosynthesis.
Plos One, 8, 2013
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | 分子名称: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | 著者 | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | 登録日 | 2020-04-01 | | 公開日 | 2020-04-08 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.95 Å) | | 主引用文献 | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
7BBG
 
 | | CRYSTAL STRUCTURE OF HLA-A2-WT1-RMF AND FAB 11D06 | | 分子名称: | Beta-2-microglobulin, Heavy chain of Fab fragment 11D06, Light chain of Fab fragment 11D06, ... | | 著者 | Bujotzek, A, Georges, G, Hanisch, L.J, Klein, C, Benz, J. | | 登録日 | 2020-12-17 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Targeting intracellular WT1 in AML with a novel RMF-peptide-MHC-specific T-cell bispecific antibody.
Blood, 138, 2021
|
|
7C2M
 
 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with NITD-349 | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Chimera of drug exporters of the RND superfamily-like protein and Endolysin, N-(4,4-dimethylcyclohexyl)-4,6-bis(fluoranyl)-1H-indole-2-carboxamide, ... | | 著者 | Zhang, B, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-05-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7CAF
 
 | | Mycobacterium smegmatis LpqY-SugABC complex in the pre-translocation state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7CAG
 
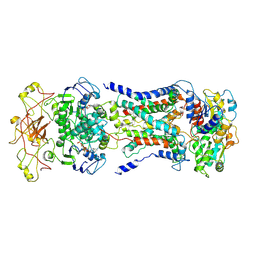 | | Mycobacterium smegmatis LpqY-SugABC complex in the catalytic intermediate state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7C2N
 
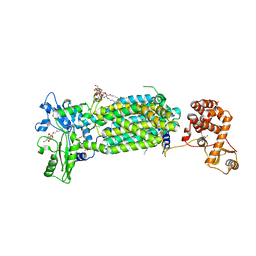 | | Crystal structure of mycolic acid transporter MmpL3 from Mycobacterium smegmatis complexed with SPIRO | | 分子名称: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, 1'-(2,3-dihydro-1,4-benzodioxin-6-ylmethyl)spiro[6,7-dihydrothieno[3,2-c]pyran-4,4'-piperidine], Drug exporters of the RND superfamily-like protein,Endolysin, ... | | 著者 | Zhang, B, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-05-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.82 Å) | | 主引用文献 | Structural Basis for the Inhibition of Mycobacterial MmpL3 by NITD-349 and SPIRO.
J.Mol.Biol., 432, 2020
|
|
7CAD
 
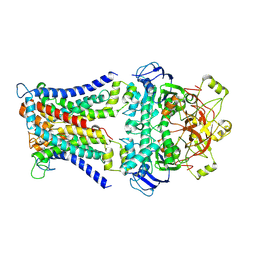 | | Mycobacterium smegmatis SugABC complex | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.41 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7CAE
 
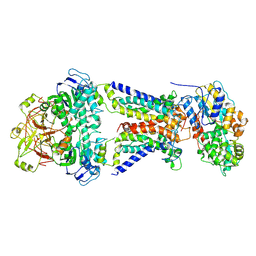 | | Mycobacterium smegmatis LpqY-SugABC complex in the resting state | | 分子名称: | ABC sugar transporter, permease component, ABC transporter, ... | | 著者 | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | 登録日 | 2020-06-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.44 Å) | | 主引用文献 | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7C53
 
 | |
3Q4I
 
 | | Crystal structure of CDP-Chase in complex with Gd3+ | | 分子名称: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | 著者 | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | 登録日 | 2010-12-23 | | 公開日 | 2011-07-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
3Q1P
 
 | | Crystal structure of CDP-Chase | | 分子名称: | Phosphohydrolase (MutT/nudix family protein), SULFATE ION | | 著者 | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | 登録日 | 2010-12-17 | | 公開日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
7XMR
 
 | | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | 分子名称: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | 著者 | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-28 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMT
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | | 分子名称: | (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | 著者 | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-28 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XMS
 
 | | CryoEM structure of somatostatin receptor 4 (SSTR4) in complex with Gi1 and its endogeneous ligand SST-14 | | 分子名称: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | 著者 | Wenli, Z, Shuo, H, Na, Q, Wenbo, Z, Mengjie, L, Dehua, Y, Ming-Wei, W, Wu, B, Zhao, Q. | | 登録日 | 2022-04-26 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
3TPE
 
 | | The phipa p3121 structure | | 分子名称: | Serine/threonine-protein kinase HipA | | 著者 | Schumacher, M.A, Link, T, Brennan, R.G. | | 登録日 | 2011-09-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2018-03-07 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3TPD
 
 | | Structure of pHipA, monoclinic form | | 分子名称: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | 著者 | schumacher, M.A, link, T, Brennan, R.G. | | 登録日 | 2011-09-07 | | 公開日 | 2012-10-03 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
