3II5
 
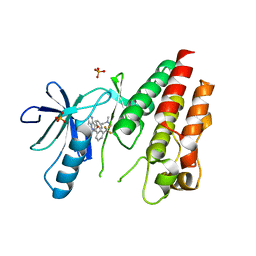 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
2HB1
 
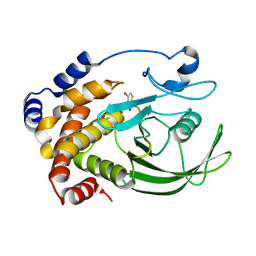 | | Crystal Structure of PTP1B with Monocyclic Thiophene Inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K, Lee, J. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2H4G
 
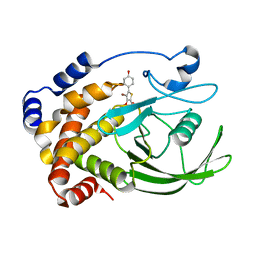 | | Crystal structure of PTP1B with monocyclic thiophene inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-(4-HYDROXYPHENYL)THIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K. | | Deposit date: | 2006-05-24 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2H4K
 
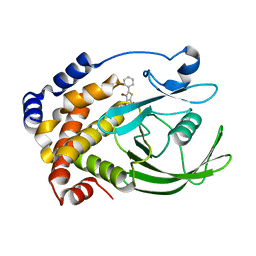 | | Crystal structure of PTP1B with a monocyclic thiophene inhibitor | | Descriptor: | 4-BROMO-3-(CARBOXYMETHOXY)-5-PHENYLTHIOPHENE-2-CARBOXYLIC ACID, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Wan, Z.-K. | | Deposit date: | 2006-05-24 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monocyclic thiophenes as protein tyrosine phosphatase 1B inhibitors: Capturing interactions with Asp48.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4M0Q
 
 | | Ebola virus VP24 structure | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Xu, W, Leung, D.W, Borek, D, Amarasinghe, G.K. | | Deposit date: | 2013-08-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | The Marburg Virus VP24 Protein Interacts with Keap1 to Activate the Cytoprotective Antioxidant Response Pathway.
Cell Rep, 6, 2014
|
|
2NTA
 
 | | Crystal Structure of PTP1B-inhibitor Complex | | Descriptor: | 5-(4-CHLORO-5-PHENYL-3-THIENYL)-1,2,5-THIADIAZOLIDIN-3-ONE 1,1-DIOXIDE, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2NT7
 
 | | Crystal structure of PTP1B-inhibitor complex | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, {[5-(3-{[1-(BENZYLSULFONYL)PIPERIDIN-4-YL]AMINO}PHENYL)-4-BROMO-2-(2H-TETRAZOL-5-YL)-3-THIENYL]OXY}ACETIC ACID | | Authors: | Xu, W, Follows, B. | | Deposit date: | 2006-11-07 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing acid replacements of thiophene PTP1B inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4FLK
 
 | | Human MetAP1 with bengamide analog Y10, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2,3-dihydro-1H-inden-2-yl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4FLI
 
 | | Human MetAP1 with bengamide analog Y16, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-(2-azanyl-2-oxidanylidene-ethyl)-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4FLJ
 
 | | Human MetAP1 with bengamide analog Y08, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-[[(3S)-1-cyclopropylcarbonylpiperidin-3-yl]methyl]-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
4FLL
 
 | | Human MetAP1 with bengamide analog YZ6, in Mn form | | Descriptor: | (E,2R,3R,4S,5R)-N-[(3R)-3-(furan-2-yl)-3-phenyl-propyl]-2-methoxy-8,8-dimethyl-3,4,5-tris(oxidanyl)non-6-enamide, MANGANESE (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Ye, Q.Z, Xu, W. | | Deposit date: | 2012-06-14 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of bengamide derivatives as inhibitors of methionine aminopeptidases.
J.Med.Chem., 55, 2012
|
|
7WHS
 
 | |
7WHR
 
 | |
7WHT
 
 | |
7V4L
 
 | |
7V4J
 
 | |
7V4H
 
 | |
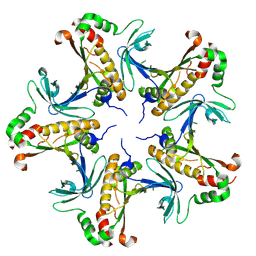
7V4I
 
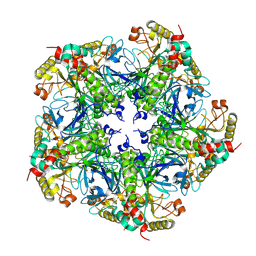 | |
7V4K
 
 | |
3KO1
 
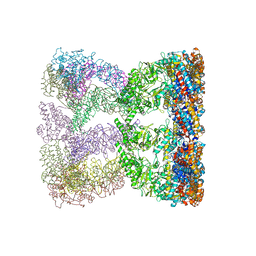 | | Cystal structure of thermosome from Acidianus tengchongensis strain S5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin | | Authors: | Huo, Y, Zhang, K, Hu, Z, Wang, L, Zhai, Y, Zhou, Q, Lander, G, He, Y, Zhu, J, Xu, W, Dong, Z, Sun, F. | | Deposit date: | 2009-11-12 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of group II chaperonin in the open state.
Structure, 18, 2010
|
|
1LUJ
 
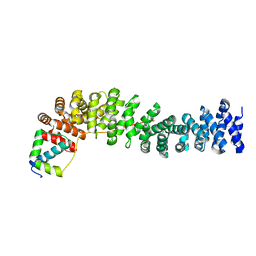 | | Crystal Structure of the Beta-catenin/ICAT Complex | | Descriptor: | Beta-catenin-interacting protein 1, Catenin beta-1 | | Authors: | Graham, T.A, Clements, W.K, Kimelman, D, Xu, W. | | Deposit date: | 2002-05-22 | | Release date: | 2002-10-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of the beta-catenin/ICAT complex reveals the inhibitory mechanism of ICAT.
Mol.Cell, 10, 2002
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
4EFB
 
 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
4FC2
 
 | |
7N7V
 
 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
