8Z8N
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 3 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z8X
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription initiation conformation | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z90
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z97
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription elongation conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z9H
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription elongation-reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z9Q
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.33 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
8Z9R
 
 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses
To Be Published
|
|
6YWC
 
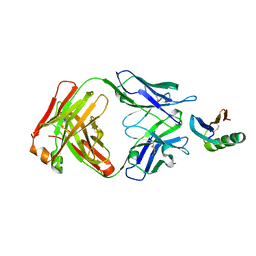 | | De novo designed protein 4E1H_95 in complex with 101F antibody | | Descriptor: | Antibody 101F, Heavy Chain, light chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
6YWD
 
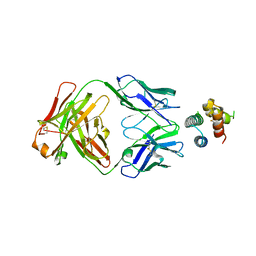 | | De novo designed protein 4H_01 in complex with Mota antibody | | Descriptor: | Antibody Mota, Heavy Chain, Light Chain, ... | | Authors: | Yang, C, Sesterhenn, F, Pojer, F, Correia, B.E. | | Deposit date: | 2020-04-29 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Bottom-up de novo design of functional proteins with complex structural features.
Nat.Chem.Biol., 17, 2021
|
|
8XJO
 
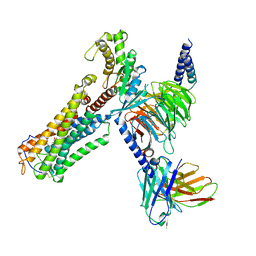 | | U46619 bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJL
 
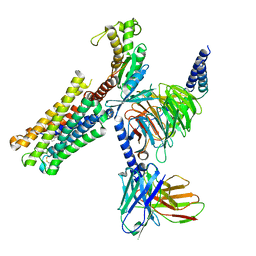 | | PGF2-alpha bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJK
 
 | | Cloprosetnol bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJN
 
 | | Cloprosetnol bound Thromboxane A2 receptor-Gq Protein Complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E},3~{R})-4-(3-chloranylphenoxy)-3-oxidanyl-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, Engineered miniGq, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
8XJM
 
 | | Latanoprost acid bound Prostaglandin F2-alpha receptor-Gq Protein Complex | | Descriptor: | Antibody fragment scFv16, Engineered miniGq, Fusion tag,Prostaglandin F2-alpha receptor,LgBiT, ... | | Authors: | Zhang, X, Li, X, Liu, G, Gong, W. | | Deposit date: | 2023-12-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for ligand recognition and activation of the prostanoid receptors.
Cell Rep, 43, 2024
|
|
4X8S
 
 | | FACTOR VIIA IN COMPLEX WITH THE INHIBITOR 4-BROMO-2-METHOXYPHENOL | | Descriptor: | 4-bromo-2-methoxyphenol, CALCIUM ION, Factor VIIa (Heavy Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-03-25 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel P1 Groups for Coagulation Factor VIIa Inhibition Using Fragment-Based Screening.
J.Med.Chem., 58, 2015
|
|
4X8U
 
 | |
4X8V
 
 | | FACTOR VIIA IN COMPLEX WITH THE INHIBITOR (methyl {3-[(2R)-1-{(2R)-2-(3,4-dimethoxyphenyl)-2-[(1-oxo-1,2,3,4-tetrahydroisoquinolin-7-yl)amino]acetyl}pyrrolidin-2-yl]-4-(propan-2-ylsulfonyl)phenyl}carbamate) | | Descriptor: | CALCIUM ION, Factor VIIa (Heavy Chain), Factor VIIa (Light Chain), ... | | Authors: | Wei, A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-03-25 | | Last modified: | 2015-04-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Novel P1 Groups for Coagulation Factor VIIa Inhibition Using Fragment-Based Screening.
J.Med.Chem., 58, 2015
|
|
4X8T
 
 | |
6S3D
 
 | |
8E5M
 
 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 6 | | Descriptor: | 1-{[(3S,4S)-3-({4-[2-(4-fluorobenzene-1-sulfonyl)ethyl]piperidin-1-yl}methyl)-4-(3-fluorophenyl)pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
8E5N
 
 | | Structure of ARG1 complex with pyrrolidine-based non-boronic acid inhibitor 10 | | Descriptor: | 1-{[(3S,4S)-3-(3-fluorophenyl)-4-{[4-(1,3,4-triethyl-1H-pyrazol-5-yl)piperidin-1-yl]methyl}pyrrolidin-1-yl]methyl}cyclopentane-1-carboxylic acid, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L, Gathiaka, S. | | Deposit date: | 2022-08-22 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Discovery of non-boronic acid Arginase 1 inhibitors through virtual screening and biophysical methods.
Bioorg.Med.Chem.Lett., 84, 2023
|
|
3I6D
 
 | | Crystal structure of PPO from bacillus subtilis with AF | | Descriptor: | 5-[2-CHLORO-4-(TRIFLUOROMETHYL)PHENOXY]-2-NITROBENZOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Shen, Y. | | Deposit date: | 2009-07-06 | | Release date: | 2009-12-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into unique properties of protoporphyrinogen oxidase from Bacillus subtilis
J.Struct.Biol., 170, 2010
|
|
6VTW
 
 | |
6XWI
 
 | |
