6XQ1
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ6
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 11 | | Descriptor: | 3-phenoxyphenol, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ3
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-(3-{[3-(4-carboxyphenoxy)phenyl]methoxy}phenyl)-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ8
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 11 | | Descriptor: | 3-phenoxyphenol, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ5
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 1 | | Descriptor: | 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XQ7
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragment 5 | | Descriptor: | 5-bromo-3-methyl-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
3CLN
 
 | |
1SAN
 
 | |
2CNP
 
 | |
1D1O
 
 | |
3PMG
 
 | |
3F7I
 
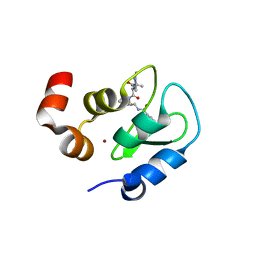 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2008-11-09 | | Release date: | 2009-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold
J.Med.Chem., 52, 2009
|
|
3F7H
 
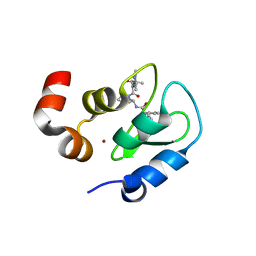 | | Structure of an ML-IAP/XIAP chimera bound to a peptidomimetic | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Baculoviral IAP repeat-containing protein 7, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2008-11-09 | | Release date: | 2009-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold
J.Med.Chem., 52, 2009
|
|
3F7G
 
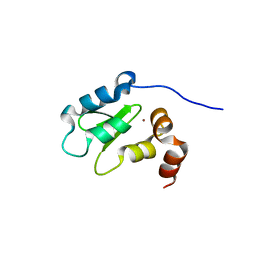 | | Structure of the BIR domain from ML-IAP bound to a peptidomimetic | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Baculoviral IAP repeat-containing protein 7, L-alanyl-L-valyl-N-(2,2-diphenylethyl)-L-prolinamide, ... | | Authors: | Franklin, M.C, Fairbrother, W.J, Cohen, F. | | Deposit date: | 2008-11-09 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Orally bioavailable antagonists of inhibitor of apoptosis proteins based on an azabicyclooctane scaffold
J.Med.Chem., 52, 2009
|
|
1CDN
 
 | |
8UCU
 
 | |
8V6I
 
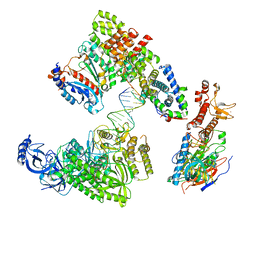 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 2024
|
|
8V5N
 
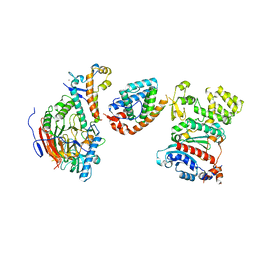 | |
8V6G
 
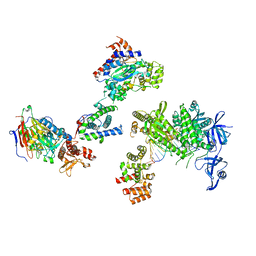 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 2024
|
|
8V6J
 
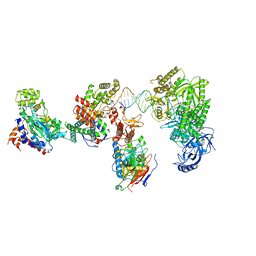 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 2024
|
|
8V5O
 
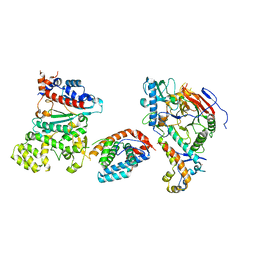 | |
8V5M
 
 | |
8V6H
 
 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 2024
|
|
1CLB
 
 | |
1FTZ
 
 | | NUCLEAR MAGNETIC RESONANCE SOLUTION STRUCTURE OF THE FUSHI TARAZU HOMEODOMAIN FROM DROSOPHILA AND COMPARISON WITH THE ANTENNAPEDIA HOMEODOMAIN | | Descriptor: | FUSHI TARAZU PROTEIN | | Authors: | Qian, Y.Q, Furukubo-Tokunaga, K, Resendez-Perez, D, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1994-01-07 | | Release date: | 1994-05-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain.
J.Mol.Biol., 238, 1994
|
|
