1XXQ
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus nigra | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
4QIA
 
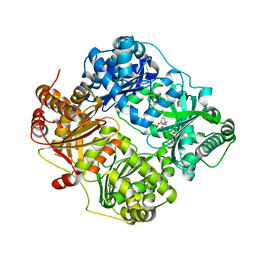 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
4R5U
 
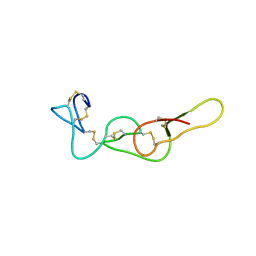 | |
4RE9
 
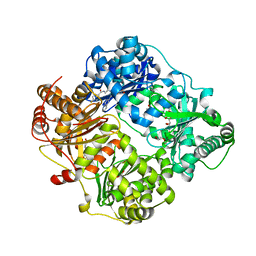 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4R4I
 
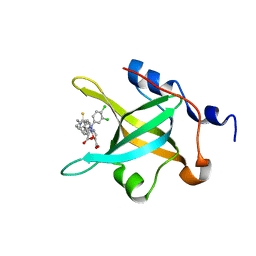 | | Structure of RPA70N in complex with 5-(4-((6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[6-(5-carboxyfuran-2-yl)-1-thioxo-3,4-dihydroisoquinolin-2(1H)-yl]methyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4O
 
 | | Crystal structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)benzyl)carbamothioyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]carbamothioyl}phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4T
 
 | | Crystal Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)phenylthioamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-{4-[({[4-(5-carboxyfuran-2-yl)phenyl]carbonothioyl}amino)methyl]phenyl}-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R4Q
 
 | | Crystal structure of RPA70N in complex with 5-(3-((N-(4-(5-carboxyfuran-2-yl)benzyl)acetamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({acetyl[4-(5-carboxyfuran-2-yl)benzyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
7TL3
 
 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF431 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.066 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL2
 
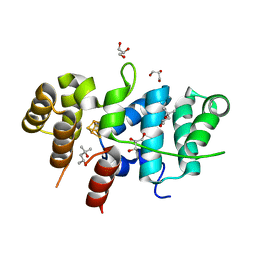 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 5YF412 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, GLYCEROL, ... | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
7TL4
 
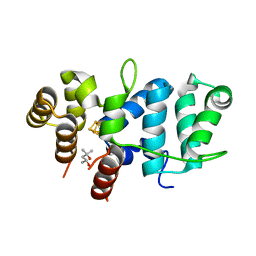 | | Crystal Structure of Yeast p58C Multi-Tyrosine Mutant 6YF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA primase large subunit, IRON/SULFUR CLUSTER | | Authors: | Blee, A.M, Salay, L.E, Chazin, W.J. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Modification of the 4Fe-4S Cluster Charge Transport Pathway Alters RNA Synthesis by Yeast DNA Primase.
Biochemistry, 61, 2022
|
|
4R4C
 
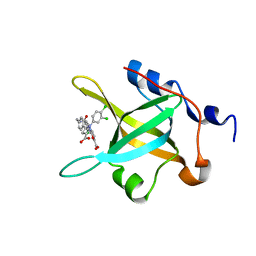 | | Structure of RPA70N in complex with 5-(4-((4-(5-carboxyfuran-2-yl)-2-chlorobenzamido)methyl)phenyl)-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid | | Descriptor: | 5-[4-({[4-(5-carboxyfuran-2-yl)-2-chlorobenzoyl]amino}methyl)phenyl]-1-(3,4-dichlorophenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Waterson, A.G, Kennedy, J.P, Patrone, J.D, Pelz, N.F, Frank, A.O, Vangamudi, B, Sousa-Fagundes, E.M, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diphenylpyrazoles as replication protein a inhibitors.
ACS Med Chem Lett, 6, 2015
|
|
4R5R
 
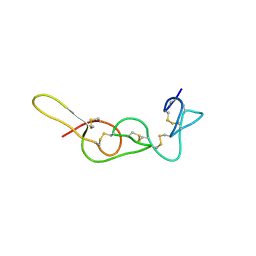 | |
4REE
 
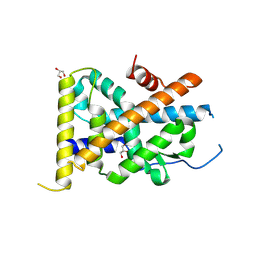 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4RA8
 
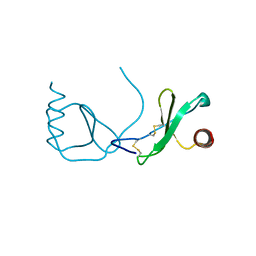 | | Structure analysis of the Mip1a P8A mutant | | Descriptor: | C-C motif chemokine 3 | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4RAL
 
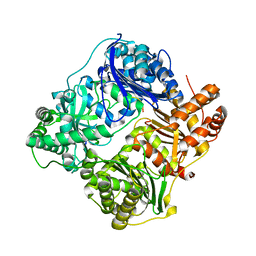 | | Crystal structure of insulin degrading enzyme in complex with macrophage inflammatory protein 1 beta | | Descriptor: | C-C motif chemokine 4, Insulin-degrading enzyme, ZINC ION | | Authors: | Liang, W.G, Ren, M, Guo, Q, Tang, W.J. | | Deposit date: | 2014-09-10 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.148 Å) | | Cite: | Structures of human CCL18, CCL3, and CCL4 reveal molecular determinants for quaternary structures and sensitivity to insulin-degrading enzyme.
J.Mol.Biol., 427, 2015
|
|
4REF
 
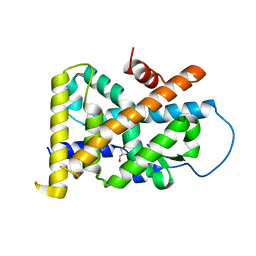 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4RPU
 
 | | Crystal Structure of Human Presequence Protease in Complex with Inhibitor MitoBloCK-60 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Mo, S.M, Liang, W.G, King, J.V, Wijaya, J, Koehler, C.M, Tang, W.J. | | Deposit date: | 2014-10-31 | | Release date: | 2015-12-09 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Crystal Structure of Human Presequence Protease in Complex with Inhibitor MitoBloCK-60
TO BE PUBLISHED
|
|
4RQG
 
 | |
4TSS
 
 | | TOXIC SHOCK SYNDROME TOXIN-1: TETRAGONAL P4(1)2(1)2 CRYSTAL FORM | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1, ZINC ION | | Authors: | Prasad, G.S, Radhakrishnan, R, Mitchell, D.T, Earhart, C.A, Dinges, M.M, Cook, W.J, Schlivert, P.M, Ohlendorf, D.H. | | Deposit date: | 1996-12-11 | | Release date: | 1997-12-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Refined structures of three crystal forms of toxic shock syndrome toxin-1 and of a tetramutant with reduced activity.
Protein Sci., 6, 1997
|
|
4U9D
 
 | |
4U5M
 
 | | Structure of a left-handed DNA G-quadruplex | | Descriptor: | DNA (28-MER), MAGNESIUM ION, POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Brahim, H, Chung, W.J, Lim, K.W. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a left-handed DNA G-quadruplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4U9E
 
 | |
4UF6
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to an activating fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, POLYUBIQUITIN-B, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UEL
 
 | | UCH-L5 in complex with ubiquitin-propargyl bound to the RPN13 DEUBAD domain | | Descriptor: | POLYUBIQUITIN-B, PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
