2AMI
 
 | | Solution Structure Of The Calcium-loaded N-Terminal Sensor Domain Of Centrin | | Descriptor: | Caltractin | | Authors: | Hu, H.T, Fagan, P.A, Bunick, C.G, Sheehan, J.H, Chazin, W.J. | | Deposit date: | 2005-08-09 | | Release date: | 2005-08-23 | | Last modified: | 2014-01-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal calcium sensor domain of centrin reveals the biochemical basis for domain-specific function.
J.Biol.Chem., 281, 2006
|
|
2B0Y
 
 | | Solution Structure of a peptide mimetic of the fourth cytoplasmic loop of the G-protein coupled CB1 cannabinoid receptor | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Grace, C.R, Cowsik, S.M, Shim, J.Y, Welsh, W.J, Howlett, A.C. | | Deposit date: | 2005-09-15 | | Release date: | 2006-08-29 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | Unique helical conformation of the fourth cytoplasmic loop of the CB1 cannabinoid receptor in a negatively charged environment.
J.Struct.Biol., 159, 2007
|
|
2BAY
 
 | | Crystal structure of the Prp19 U-box dimer | | Descriptor: | Pre-mRNA splicing factor PRP19 | | Authors: | Vander Kooi, C.W, Ohi, M.D, Rosenberg, J.A, Oldham, M.L, Newcomer, M.E, Gould, K.L, Chazin, W.J. | | Deposit date: | 2005-10-15 | | Release date: | 2006-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Prp19 U-box Crystal Structure Suggests a Common Dimeric Architecture for a Class of Oligomeric E3 Ubiquitin Ligases.
Biochemistry, 45, 2006
|
|
2BCT
 
 | |
1ULA
 
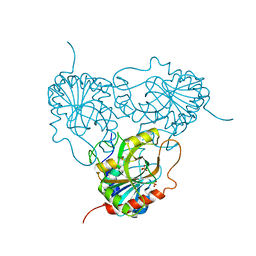 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1VCJ
 
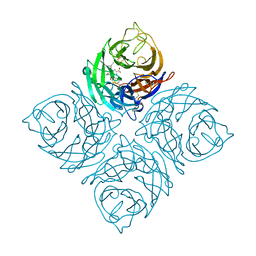 | | Influenza B virus neuraminidase complexed with 1-(4-Carboxy-2-(3-pentylamino)phenyl)-5-aminomethyl-5-hydroxymethyl-pyrrolidin-2-one | | Descriptor: | 4-[(2R)-2-(AMINOMETHYL)-2-(HYDROXYMETHYL)-5-OXOPYRROLIDIN-1-YL]-3-[(1-ETHYLPROPYL)AMINO]BENZOIC ACID, NEURAMINIDASE | | Authors: | Lommer, B.S, Ali, S.M, Bajpai, S.N, Brouillette, W.J, Air, G.M, Luo, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A benzoic acid inhibitor induces a novel conformational change in the active site of Influenza B virus neuraminidase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1VSU
 
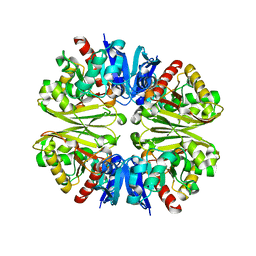 | |
2BEP
 
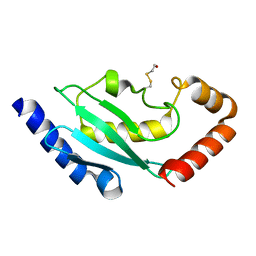 | | Crystal structure of ubiquitin conjugating enzyme E2-25K | | Descriptor: | BETA-MERCAPTOETHANOL, UBIQUITIN-CONJUGATING ENZYME E2-25 KDA | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-11-29 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
1VGH
 
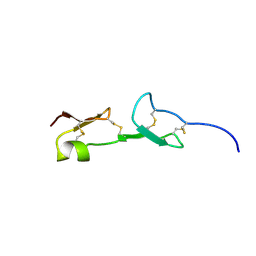 | | HEPARIN-BINDING DOMAIN FROM VASCULAR ENDOTHELIAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR-165 | | Authors: | Fairbrother, W.J, Champe, M.A, Christinger, H.W, Keyt, B.A, Starovasnik, M.A. | | Deposit date: | 1997-12-17 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heparin-binding domain of vascular endothelial growth factor.
Structure, 6, 1998
|
|
1VSV
 
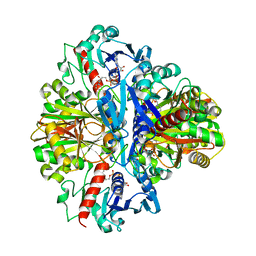 | |
1VPP
 
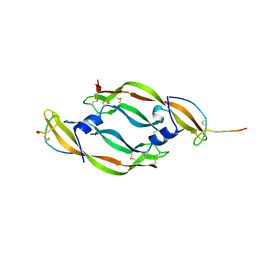 | | COMPLEX BETWEEN VEGF AND A RECEPTOR BLOCKING PEPTIDE | | Descriptor: | PROTEIN (PEPTIDE V108), PROTEIN (VASCULAR ENDOTHELIAL GROWTH FACTOR) | | Authors: | Wiesmann, C, Christinger, H.W, Cochran, A.G, Cunningham, B.C, Fairbrother, W.J, Keenan, C.J, Meng, G, de Vos, A.M. | | Deposit date: | 1998-10-09 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between VEGF and a receptor-blocking peptide.
Biochemistry, 37, 1998
|
|
2BF8
 
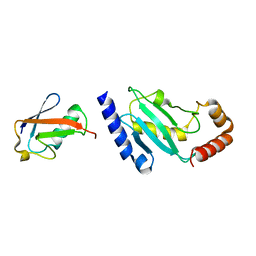 | | Crystal structure of SUMO modified ubiquitin conjugating enzyme E2- 25K | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-25 KDA, UBIQUITIN-LIKE PROTEIN SMT3C | | Authors: | Pichler, A, Knipscheer, P, Oberhofer, E, Van Dijk, W.J, Korner, R, Velgaard Olsen, J, Jentsch, S, Melchior, F, Sixma, T.K. | | Deposit date: | 2004-12-06 | | Release date: | 2005-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sumo Imodification of the Ubiquitin Conjugating Enzyme E2-25K
Nat.Struct.Mol.Biol., 12, 2005
|
|
2CYG
 
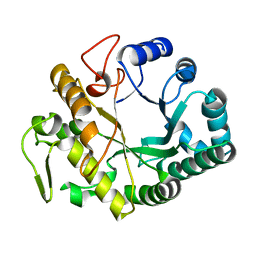 | | Crystal structure at 1.45- resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome | | Descriptor: | beta-1, 3-glucananse | | Authors: | Receveur-Brechot, V, Czjzek, M, Barre, A, Roussel, A, Peumans, W.J, Van Damme, E.J.M, Rouge, P. | | Deposit date: | 2005-07-07 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure at 1.45-A resolution of the major allergen endo-beta-1,3-glucanase of banana as a molecular basis for the latex-fruit syndrome
Proteins, 63, 2006
|
|
2COL
 
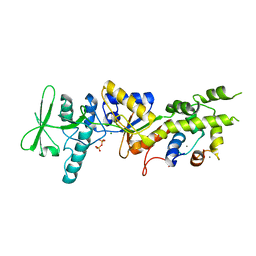 | | Crystal structure analysis of CyaA/C-Cam with Pyrophosphate | | Descriptor: | Bifunctional hemolysin-adenylate cyclase, CALCIUM ION, Calmodulin, ... | | Authors: | Guo, Q, Tang, W.J, Shen, Y. | | Deposit date: | 2005-05-18 | | Release date: | 2006-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin
Embo J., 24, 2005
|
|
1XFX
 
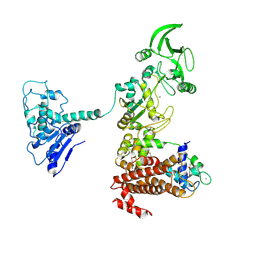 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 10 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2017-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFZ
 
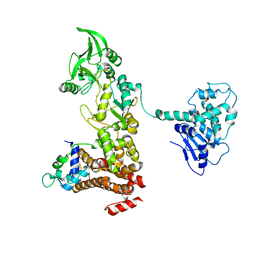 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin in the presence of 1 millimolar exogenously added calcium chloride | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
2D82
 
 | | Target Structure-Based Discovery of Small Molecules that Block Human p53 and CREB Binding Protein (CBP) Association | | Descriptor: | 9-ACETYL-2,3,4,9-TETRAHYDRO-1H-CARBAZOL-1-ONE, CREB-binding protein | | Authors: | Sachchidanand, Resnick-Silverman, L, Yan, S, Mujtaba, S, Liu, W.J, Zeng, L, Manfredi, J.J, Zhou, M.M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-04-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Target structure-based discovery of small molecules that block human p53 and CREB binding protein association
Chem.Biol., 13, 2006
|
|
1XFU
 
 | | Crystal structure of anthrax edema factor (EF) truncation mutant, EF-delta 64 in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
1XFY
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin | | Descriptor: | CALCIUM ION, Calmodulin 2, Calmodulin-sensitive adenylate cyclase, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFW
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3'5' cyclic AMP (cAMP) | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Y, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XFV
 
 | | Crystal structure of anthrax edema factor (EF) in complex with calmodulin and 3' deoxy-ATP | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, Calmodulin 2, ... | | Authors: | Shen, Q, Zhukovskaya, N.L, Guo, Q, Florian, J, Tang, W.J. | | Deposit date: | 2004-09-15 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor.
EMBO J., 24, 2005
|
|
1XMN
 
 | | Crystal structure of thrombin bound to heparin | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ... | | Authors: | Carter, W.J, Cama, E, Huntington, J.A. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of thrombin bound to heparin
J.Biol.Chem., 280, 2005
|
|
1XXR
 
 | | Structure of a mannose-specific jacalin-related lectin from Morus Nigra in complex with mannose | | Descriptor: | ACETIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Rabijns, A, Barre, A, Van Damme, E.J.M, Peumans, W.J, De Ranter, C.J, Rouge, P. | | Deposit date: | 2004-11-08 | | Release date: | 2005-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of the jacalin-related lectin MornigaM from the black mulberry (Morus nigra) in complex with mannose
Febs J., 272, 2005
|
|
