1SYQ
 
 | |
1TR2
 
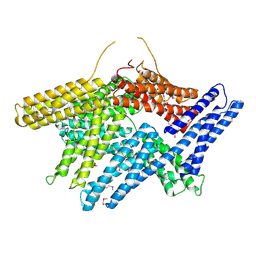 | | Crystal structure of human full-length vinculin (residues 1-1066) | | Descriptor: | VINCULIN ISOFORM 1 | | Authors: | Borgon, R.A, Vonrhein, C, Bricogne, G, Bois, P.R, Izard, T. | | Deposit date: | 2004-06-19 | | Release date: | 2005-11-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Human Vinculin
Structure, 12, 2004
|
|
4AWL
 
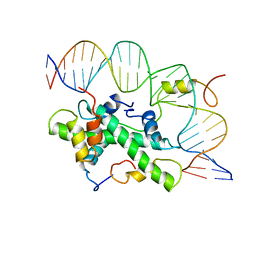 | | The NF-Y transcription factor is structurally and functionally a sequence specific histone | | Descriptor: | HSP70 PROMOTER FRAGMENT, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT ALPHA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, ... | | Authors: | Nardini, M, Gnesutta, N, Donati, G, Gatta, R, Forni, C, Fossati, A, Vonrhein, C, Moras, D, Romier, C, Mantovani, R, Bolognesi, M. | | Deposit date: | 2012-06-04 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Sequence-Specific Transcription Factor NF-Y Displays Histone-Like DNA Binding and H2B-Like Ubiquitination.
Cell(Cambridge,Mass.), 152, 2013
|
|
1W0E
 
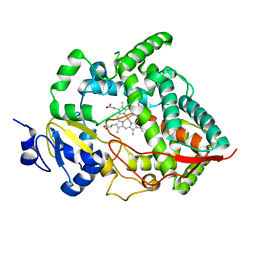 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
1W0G
 
 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, METYRAPONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
1W0F
 
 | | Crystal structure of human cytochrome P450 3A4 | | Descriptor: | CYTOCHROME P450 3A4, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Williams, P.A, Cosme, J, Vinkovic, D.M, Ward, A, Angove, H.C, Day, P.J, Vonrhein, C, Tickle, I.J, Jhoti, H. | | Deposit date: | 2004-06-03 | | Release date: | 2004-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structures of Human Cytochrome P450 3A4 Bound to Metyrapone and Progesterone
Science, 305, 2004
|
|
2WJ8
 
 | | Respiratory Syncitial Virus RiboNucleoProtein | | Descriptor: | BORATE ION, NUCLEOPROTEIN, RNA (5'-R(*CP*CP*CP*CP*CP*C)-3') | | Authors: | Tawar, R.G, Duquerroy, S, Vonrhein, C, Varela, P.F, Damier-Piolle, L, Castagne, N, MacLellan, K, Bedouelle, H, Bricogne, G, Bhella, D, Eleouet, J, Rey, F.A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-12-08 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal Structure of a Nucleocapsid-Like Nucleoprotein-RNA Complex of Respiratory Syncytial Virus
Science, 326, 2009
|
|
2F5I
 
 | |
1J5E
 
 | | Structure of the Thermus thermophilus 30S Ribosomal Subunit | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Wimberly, B.T, Brodersen, D.E, Clemons Jr, W.M, Morgan-Warren, R, Carter, A.P, Vonrhein, C, Hartsch, T, Ramakrishnan, V. | | Deposit date: | 2002-04-08 | | Release date: | 2002-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the 30S ribosomal subunit.
Nature, 407, 2000
|
|
1LF1
 
 | | Crystal Structure of Cel5 from Alkalophilic Bacillus sp. | | Descriptor: | Cel5 | | Authors: | Shaw, A, Bott, R, Vonrhein, C, Bricogne, G, Power, S, Day, A.G. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A novel combination of two classic catalytic schemes.
J.Mol.Biol., 320, 2002
|
|
1MAW
 
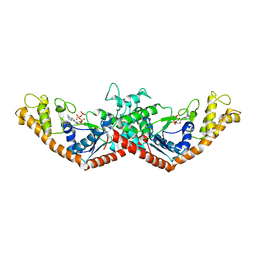 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in an Open Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1M83
 
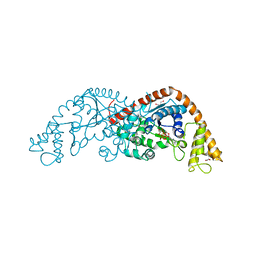 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with ATP in a Closed, Pre-transition State Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-07-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MAU
 
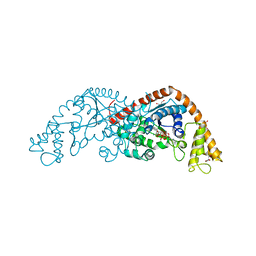 | | Crystal structure of Tryptophanyl-tRNA Synthetase Complexed with ATP and Tryptophanamide in a Pre-Transition state Conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CITRIC ACID, GLYCEROL, ... | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
1MB2
 
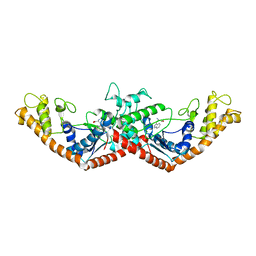 | | Crystal Structure of Tryptophanyl-tRNA Synthetase Complexed with Tryptophan in an Open Conformation | | Descriptor: | TRYPTOPHAN, TRYPTOPHAN-TRNA LIGASE | | Authors: | Retailleau, P, Huang, X, Yin, Y, Hu, M, Weinreb, V, Vachette, P, Vonrhein, C, Bricogne, G, Roversi, P, Ilyin, V, Carter Jr, C.W. | | Deposit date: | 2002-08-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interconversion of ATP binding and conformational free energies by tryptophanyl-tRNA synthetase: structures of ATP bound to open and closed, pre-transition-state conformations.
J.Mol.Biol., 325, 2003
|
|
2VKZ
 
 | | Structure of the cerulenin-inhibited fungal fatty acid synthase type I multienzyme complex | | Descriptor: | (2S, 3R)-3-HYDROXY-4-OXO-7,10-TRANS,TRANS-DODECADIENAMIDE, FATTY ACID SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Johansson, P, Wiltschi, B, Kumari, P, Kessler, B, Vonrhein, C, Vonck, J, Oesterhelt, D, Grininger, M. | | Deposit date: | 2008-01-07 | | Release date: | 2008-08-12 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Inhibition of the Fungal Fatty Acid Synthase Type I Multienzyme Complex.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
2WCT
 
 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
2XMH
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CITRATE ANION, CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Archaeoglobus fulgidus CTP:inositol-1-phosphate cytidylyltransferase, a key enzyme for di-myo-inositol-phosphate synthesis in (hyper)thermophiles.
J. Bacteriol., 193, 2011
|
|
2XME
 
 | | The X-ray structure of CTP:inositol-1-phosphate cytidylyltransferase from Archaeoglobus fulgidus | | Descriptor: | CTP-INOSITOL-1-PHOSPHATE CYTIDYLYLTRANSFERASE, GLYCEROL | | Authors: | Brito, J.A, Borges, N, Vonrhein, C, Santos, H, Archer, M. | | Deposit date: | 2010-07-27 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Archaeoglobus Fulgidus Ctp:Inositol-1-Phosphate Cytidylyltransferase, a Key Enzyme for Di-Myo-Inositol-Phosphate Synthesis in (Hyper)Thermophiles.
J.Bacteriol., 193, 2011
|
|
2XSZ
 
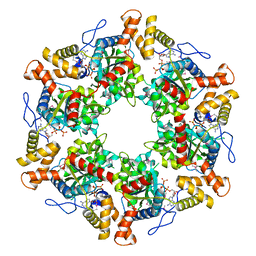 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
2YLE
 
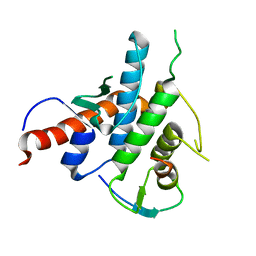 | | Crystal structure of the human Spir-1 KIND FSI domain in complex with the FSI peptide | | Descriptor: | FORMIN-2, PROTEIN SPIRE HOMOLOG 1 | | Authors: | Zeth, K, Pechlivanis, M, Vonrhein, C, Kerkhoff, E. | | Deposit date: | 2011-06-01 | | Release date: | 2011-06-08 | | Last modified: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Basis of Actin Nucleation Factor Cooperativity: Crystal Structure of the Spir-1 Kinase Non-Catalytic C-Lobe Domain (Kind)Formin-2 Formin Spir Interaction Motif (Fsi) Complex.
J.Biol.Chem., 286, 2011
|
|
2YLF
 
 | |
4CBG
 
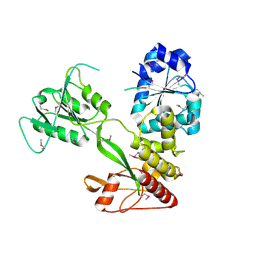 | | Pestivirus NS3 helicase | | Descriptor: | ACETATE ION, SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBM
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
4CBL
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
