4X2M
 
 | | Structure of Mtr2 | | Descriptor: | Mtr2 | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of the principal mRNA-export factor Mex67-Mtr2 from Chaetomium thermophilum.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4X2H
 
 | | Sac3N peptide bound to Mex67:Mtr2 | | Descriptor: | Putative mRNA export protein, Putative uncharacterized protein, SER-SER-VAL-PHE-GLY-ALA-PRO-ALA | | Authors: | Aibara, S, Valkov, E, Stewart, M. | | Deposit date: | 2014-11-26 | | Release date: | 2015-06-17 | | Last modified: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2.
Structure, 23, 2015
|
|
5NVN
 
 | | Crystal structure of the human 4EHP-4E-BP1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, Eukaryotic translation initiation factor 4E-binding protein 1, FORMIC ACID | | Authors: | Peter, D, Sandmeir, F, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5NVM
 
 | |
5NVK
 
 | | Crystal structure of the human 4EHP-GIGYF1 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 1 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
5NVL
 
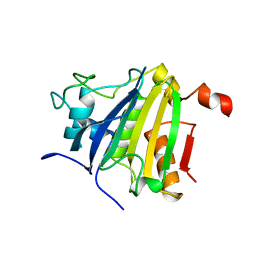 | | Crystal structure of the human 4EHP-GIGYF2 complex | | Descriptor: | Eukaryotic translation initiation factor 4E type 2, GRB10-interacting GYF protein 2 | | Authors: | Peter, D, Valkov, E. | | Deposit date: | 2017-05-04 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.
Genes Dev., 31, 2017
|
|
3OZI
 
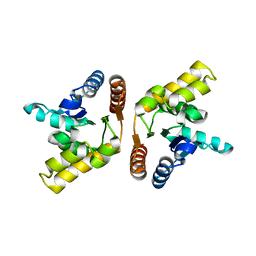 | | Crystal structure of the TIR domain from the flax disease resistance protein L6 | | Descriptor: | COBALT (II) ION, L6tr | | Authors: | Ve, T, Bernoux, M, Williams, S, Valkov, E, Warren, C, Hatters, D, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2010-09-25 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of a Plant Resistance Protein TIR Domain Reveals Interfaces for Self-Association, Signaling, and Autoregulation.
Cell Host Microbe, 9, 2011
|
|
3PSN
 
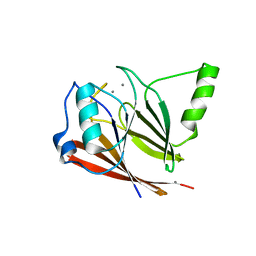 | | Crystal structure of mouse VPS29 complexed with Mn2+ | | Descriptor: | MANGANESE (II) ION, Vacuolar protein sorting-associated protein 29 | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
3PSO
 
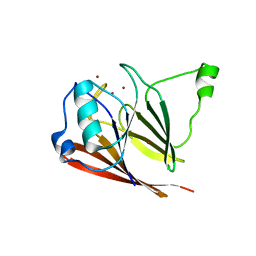 | | Crystal structure of mouse VPS29 complexed with Zn2+ | | Descriptor: | Vacuolar protein sorting-associated protein 29, ZINC ION | | Authors: | Swarbrick, J, Shaw, D, Chhabra, S, Ghai, R, Valkov, E, Norwood, S, Collins, B. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational dynamics and biomolecular interactions of VPS29 studied by NMR and X-ray crystallography
To be Published
|
|
6H3Z
 
 | | Crystal structure of a C-terminal MIF4G domain in NOT1 | | Descriptor: | CCR4-not transcription complex subunit 1, SODIUM ION | | Authors: | Raisch, T, Sandmeir, F, Weichenrieder, O, Valkov, E, Izaurralde, E. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4-NOT complex.
J. Struct. Biol., 204, 2018
|
|
6FC2
 
 | |
6FBZ
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum in the cap-bound state | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-like protein,Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6FC3
 
 | |
6FC1
 
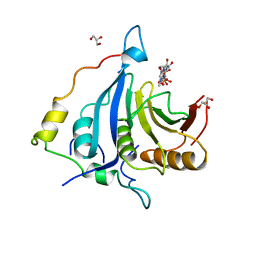 | |
6FC0
 
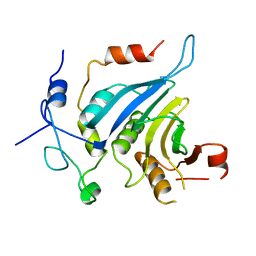 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum | | Descriptor: | Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
4WYK
 
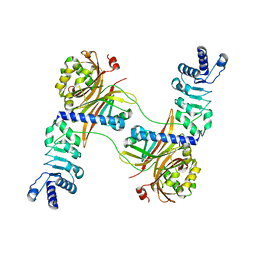 | |
4U5L
 
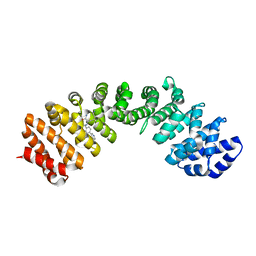 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5V
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-{[5-(pyridin-3-yl)thiophen-2-yl]methyl}-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5N
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-L-lysyl-N-[(1R,2S,3R)-1-{[(2R)-1-amino-1-oxo-3-phenylpropan-2-yl]amino}-1,3-dihydroxybutan-2-yl]glycinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5S
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N-[(2S)-2-[(N~2~-acetyl-D-lysyl)amino]-3-(pyridin-3-ylmethoxy)propyl]-L-allothreonyl-D-phenylalaninamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U54
 
 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N-methyl-1-[3-(pyridin-3-yl)phenyl]methanamine | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U58
 
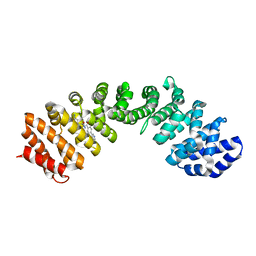 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzoyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-24 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5O
 
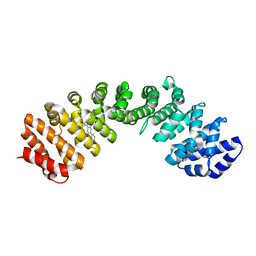 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[4-(pyridin-3-yl)benzyl]-D-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
4U5U
 
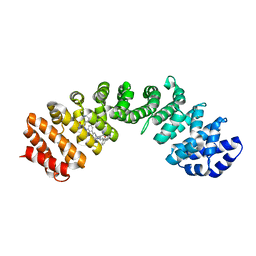 | | IMPORTIN-ALPHA MINOR NLS SITE INHIBITOR | | Descriptor: | Importin subunit alpha-1, N~2~-[3-(pyridin-3-yl)benzyl]-L-lysinamide | | Authors: | Stewart, M, Valkov, E, Holvey, R.S. | | Deposit date: | 2014-07-25 | | Release date: | 2015-05-13 | | Last modified: | 2015-07-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Selective Targeting of the TPX2 Site of Importin-alpha Using Fragment-Based Ligand Design.
Chemmedchem, 10, 2015
|
|
3ZLG
 
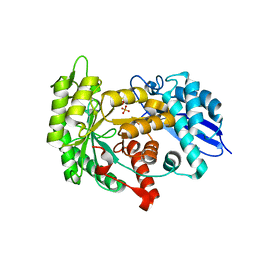 | | Structure of group A Streptococcal enolase K362A mutant | | Descriptor: | ENOLASE, PHOSPHATE ION | | Authors: | Cork, A.J, Ericsson, D.J, Law, R.H.P, Casey, L.W, Valkov, E, Bertozzi, C, Stamp, A, Aquilina, J.A, Whisstock, J.C, Walker, M.J, Kobe, B. | | Deposit date: | 2013-01-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Stability of the Octameric Structure Affects Plasminogen-Binding Capacity of Streptococcal Enolase.
Plos One, 10, 2015
|
|
