1EJE
 
 | | CRYSTAL STRUCTURE OF AN FMN-BINDING PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-BINDING PROTEIN, NICKEL (II) ION, ... | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Arrowsmith, C, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-02 | | Release date: | 2000-10-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural proteomics of an archaeon.
Nat.Struct.Biol., 7, 2000
|
|
9BHS
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-9939 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-(1-methyl-1H-pyrazol-4-yl)-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1, UNKNOWN LIGAND | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-9939 compound
To be published
|
|
9BHR
 
 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-{(1E)-3-[(2-methoxyethyl)amino]-3-oxoprop-1-en-1-yl}-1H-pyrrole-3-carboxamide, DDB1- and CUL4-associated factor 1 | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-Awar, R, Krausser, C, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-40155 compound
To be published
|
|
4U7D
 
 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | Descriptor: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1EF4
 
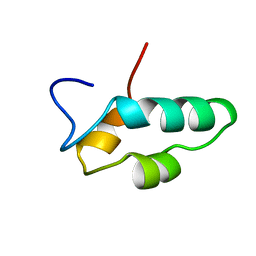 | | SOLUTION STRUCTURE OF THE ESSENTIAL RNA POLYMERASE SUBUNIT RPB10 FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DNA-DIRECTED RNA POLYMERASE, ZINC ION | | Authors: | Mackereth, C.D, Arrowsmith, C.H, Edwards, A.M, Mcintosh, L.P, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-07 | | Release date: | 2000-06-14 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EIK
 
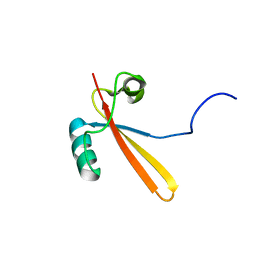 | | Solution Structure of RNA Polymerase Subunit RPB5 from Methanobacterium Thermoautotrophicum | | Descriptor: | RNA POLYMERASE SUBUNIT RPB5 | | Authors: | Yee, A, Booth, V, Dharamsi, A, Engel, A, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-02-25 | | Release date: | 2000-06-21 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RNA polymerase subunit RPB5 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EO0
 
 | | CONSERVED DOMAIN COMMON TO TRANSCRIPTION FACTORS TFIIS, ELONGIN A, CRSP70 | | Descriptor: | TRANSCRIPTION ELONGATION FACTOR S-II | | Authors: | Booth, V, Koth, C, Edwards, A.M, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-21 | | Release date: | 2000-05-03 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structure of a conserved domain common to the transcription factors TFIIS, elongin A, and CRSP70
J.Biol.Chem., 275, 2000
|
|
1DQZ
 
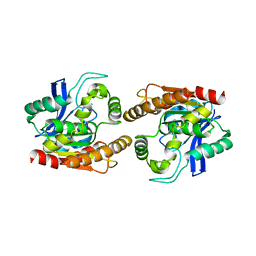 | |
1DQY
 
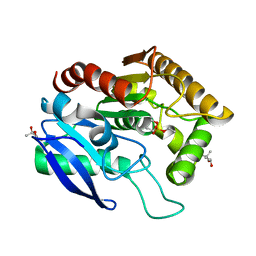 | | CRYSTAL STRUCTURE OF ANTIGEN 85C FROM MYCOBACTERIUM TUBERCULOSIS WITH DIETHYL PHOSPHATE INHIBITOR | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DIETHYL PHOSPHONATE, PROTEIN (ANTIGEN 85-C) | | Authors: | Ronning, D.R, Klabunde, T, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-01-05 | | Release date: | 2000-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of the secreted form of antigen 85C reveals potential targets for mycobacterial drugs and vaccines.
Nat.Struct.Biol., 7, 2000
|
|
1ENY
 
 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
1ENZ
 
 | | CRYSTAL STRUCTURE AND FUNCTION OF THE ISONIAZID TARGET OF MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dessen, A, Quemard, A, Blanchard, J.S, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1995-01-27 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and function of the isoniazid target of Mycobacterium tuberculosis.
Science, 267, 1995
|
|
1EP0
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | DTDP-6-DEOXY-D-XYLO-4-HEXULOSE 3,5-EPIMERASE | | Authors: | Christendat, D, Saridakis, V, Bochkarev, A, Pai, E.F, Arrowsmith, C.H, Edwards, A.M, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-03-24 | | Release date: | 2000-12-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dTDP-4-keto-6-deoxy-D-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP.
J.Biol.Chem., 275, 2000
|
|
4UAQ
 
 | | Crystal structure of the accessory translocation ATPase, SecA2, from Mycobacterium tuberculosis | | Descriptor: | Protein translocase subunit SecA 2 | | Authors: | Swanson-Smith, S, Ioerger, T.R, Rigel, N.W, Miller, B.K, Braunstein, M, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2014-08-11 | | Release date: | 2015-09-09 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Similarities and Differences between Two Functionally Distinct SecA Proteins, Mycobacterium tuberculosis SecA1 and SecA2.
J.Bacteriol., 198, 2015
|
|
1C3V
 
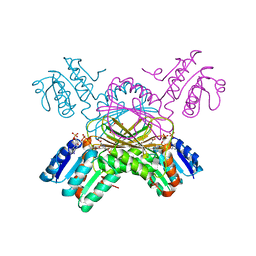 | | DIHYDRODIPICOLINATE REDUCTASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND PDC | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1999-07-28 | | Release date: | 2003-08-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis
dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes.
Structural and mutagenic analysis of relaxed nucleotide specificity
Biochemistry, 42, 2003
|
|
1BVR
 
 | | M.TB. ENOYL-ACP REDUCTASE (INHA) IN COMPLEX WITH NAD+ AND C16-FATTY-ACYL-SUBSTRATE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE), TRANS-2-HEXADECENOYL-(N-ACETYL-CYSTEAMINE)-THIOESTER | | Authors: | Rozwarski, D.A, Vilcheze, C, Sugantino, M, Bittman, R, Jacobs, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
J.Biol.Chem., 274, 1999
|
|
7KBQ
 
 | |
7FHJ
 
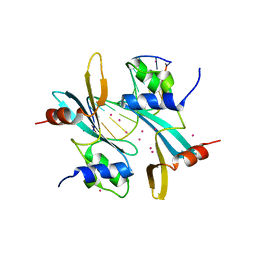 | | Crystal structure of BAZ2A with DNA | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, DNA (5'-D(*CP*GP*GP*AP*AP*TP*GP*TP*AP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*TP*AP*(5CM)P*AP*TP*TP*CP*CP*G)-3'), ... | | Authors: | Liu, K, Dong, A, Li, Y, Loppnau, P, Edwards, A.M, Arrowsmith, C.H, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of the TAM domain of BAZ2A in binding to DNA or RNA independent of methylation status.
J.Biol.Chem., 297, 2021
|
|
7TZD
 
 | |
7TZ8
 
 | |
4XYJ
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ATP and Mg, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
4XYK
 
 | | Crystal structure of human phosphofructokinase-1 in complex with ADP, Northeast Structural Genomics Consortium Target HR9275 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent 6-phosphofructokinase, platelet type, ... | | Authors: | Forouhar, F, Webb, B.A, Szu, F.-E, Seetharaman, J, Barber, D.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-02-02 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations.
Nature, 523, 2015
|
|
7B6M
 
 | | Crystal structure of MurE from E.coli in complex with Z1198948504 | | Descriptor: | 1,2-ETHANEDIOL, N-[2-(2,5-Dioxopyrrolidin-1-yl)ethyl]-3-methylbenzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC), Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
8C13
 
 | | Crystal structure of pVHL:ElonginC:ElonginB complex bound to PROTAC JW48 | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[3-[2-[2-[2-(acetamidomethyl)-4-(6,7-dihydro-5~{H}-pyrrolo[1,2-a]imidazol-2-yl)phenoxy]ethoxy]ethoxy]propanoylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Weckesser, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tracking the PROTAC degradation pathway in living cells highlights the importance of ternary complex measurement for PROTAC optimization.
Cell Chem Biol, 30, 2023
|
|
5KPE
 
 | | Solution NMR Structure of Denovo Beta Sheet Design Protein, Northeast Structural Genomics Consortium (NESG) Target OR664 | | Descriptor: | De novo Beta Sheet Design Protein OR664 | | Authors: | Tang, Y, Liu, G, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2016-07-03 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
5KPH
 
 | |
