1TVW
 
 | |
2NG1
 
 | | N AND GTPASE DOMAINS OF THE SIGNAL SEQUENCE RECOGNITION PROTEIN FFH FROM THERMUS AQUATICUS | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Freymann, D.M, Stroud, R.M, Walter, P. | | Deposit date: | 1998-09-11 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+GDP.
Nat.Struct.Biol., 6, 1999
|
|
1TSV
 
 | | THYMIDYLATE SYNTHASE R179A MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TSW
 
 | | THYMIDYLATE SYNTHASE R179A MUTANT | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1TVV
 
 | |
1TSY
 
 | | THYMIDYLATE SYNTHASE R179K MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
2TSC
 
 | | STRUCTURE, MULTIPLE SITE BINDING, AND SEGMENTAL ACCOMODATION IN THYMIDYLATE SYNTHASE ON BINDING D/UMP AND AN ANTI-FOLATE | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Montfort, W.R, Stroud, R.M. | | Deposit date: | 1991-07-03 | | Release date: | 1991-10-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure, multiple site binding, and segmental accommodation in thymidylate synthase on binding dUMP and an anti-folate.
Biochemistry, 29, 1990
|
|
2TGD
 
 | |
2EVU
 
 | | Crystal structure of aquaporin AqpM at 2.3A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-10-31 | | Release date: | 2005-12-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2F2B
 
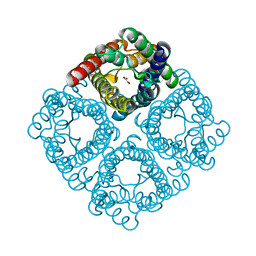 | | Crystal structure of integral membrane protein Aquaporin AqpM at 1.68A resolution | | Descriptor: | Aquaporin aqpM, GLYCEROL | | Authors: | Lee, J.K, Kozono, D, Remis, J, Kitagawa, Y, Agre, P, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2005-11-15 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
4FGT
 
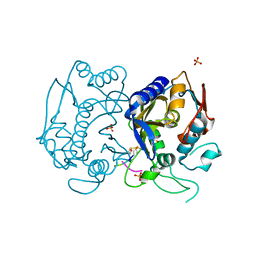 | | Allosteric peptidic inhibitor of human thymidylate synthase that stabilizes inactive conformation of the enzyme. | | Descriptor: | CG peptide, SULFATE ION, Thymidylate synthase | | Authors: | Tochowicz, A, Finer-Moore, J, Stroud, R.M, Costi, M.P. | | Deposit date: | 2012-06-04 | | Release date: | 2013-03-06 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alanine mutants of the interface residues of human thymidylate synthase decode key features of the binding mode of allosteric anticancer peptides.
J.Med.Chem., 58, 2015
|
|
3AID
 
 | |
2FR0
 
 | |
2FR1
 
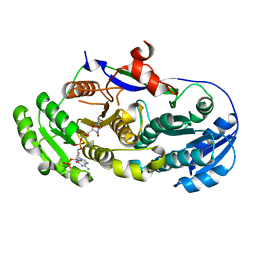 | |
3BHS
 
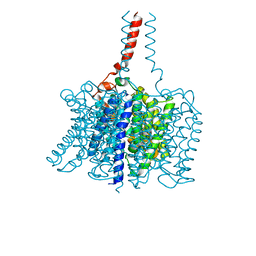 | | Nitrosomonas europaea Rh50 and mechanism of conduction by Rhesus protein family of channels | | Descriptor: | Ammonium transporter family protein Rh50 | | Authors: | Gruswitz, F, Ho, C.-M, del Rosario, M.C, Westhoff, C.M, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2007-11-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Nitrosomonas europaea Rh50 and mechanism of conduction by Rhesus protein family of channels.
To be Published
|
|
2G8A
 
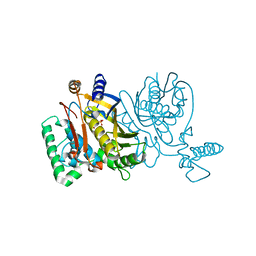 | | Lactobacillus casei Y261M in complex with substrate, dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, thymidylate synthase | | Authors: | Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-02 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2'-deoxyuridine 5'-monophosphate (dUMP)-binding Tyr-261
Biochemistry, 45, 2006
|
|
2G8X
 
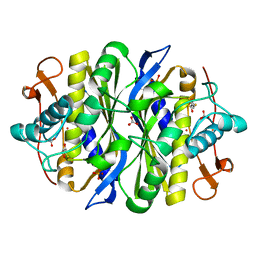 | | Escherichia coli Y209W apoprotein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Lee, T.T, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 2006-03-03 | | Release date: | 2006-03-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The role of protein dynamics in thymidylate synthase catalysis: Variants of conserved dUMP-binding Tyr-261
TO BE PUBLISHED
|
|
2G86
 
 | |
2G8D
 
 | |
2G89
 
 | |
3BT7
 
 | |
4GGM
 
 | | Structure of LpxI | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, MAGNESIUM ION, UDP-2,3-diacylglucosamine pyrophosphatase LpxI | | Authors: | Metzger IV, L.E, Lee, J.K, Finer-Moore, J.S, Raetz, C.R.H, Stroud, R.M, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-03 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | LpxI structures reveal how a lipid A precursor is synthesized.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3C02
 
 | |
3C4B
 
 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-29 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3C4T
 
 | | Structure of RNaseIIIb and dsRNA binding domains of mouse Dicer | | Descriptor: | CADMIUM ION, Endoribonuclease Dicer | | Authors: | Lee, J.K, Du, Z, Tjhen, R.J, Stroud, R.M, James, T.L. | | Deposit date: | 2008-01-30 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical insights into the dicing mechanism of mouse Dicer: A conserved lysine is critical for dsRNA cleavage.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
