1B0A
 
 | | 5,10, METHYLENE-TETRAHYDROPHOLATE DEHYDROGENASE/CYCLOHYDROLASE FROM E COLI. | | Descriptor: | PROTEIN (FOLD BIFUNCTIONAL PROTEIN) | | Authors: | Shen, B.W, Dyer, D, Huang, J.-Y, D'Ari, L, Rabinowitz, J, Stoddard, B.L. | | Deposit date: | 1998-11-06 | | Release date: | 1999-06-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of a bacterial, bifunctional 5,10 methylene-tetrahydrofolate dehydrogenase/cyclohydrolase.
Protein Sci., 8, 1999
|
|
2EX5
 
 | |
2FLD
 
 | | I-MsoI Re-Designed for Altered DNA Cleavage Specificity | | Descriptor: | 5'-D(*CP*GP*GP*AP*AP*CP*GP*GP*TP*CP*TP*CP*AP*CP*GP*AP*CP*CP*TP*TP*CP*TP*GP*C)-3', 5'-D(*GP*CP*AP*GP*AP*AP*GP*GP*TP*CP*GP*TP*GP*AP*GP*AP*CP*CP*GP*TP*TP*CP*CP*G)-3', CALCIUM ION, ... | | Authors: | Ashworth, J, Duarte, C.M, Havranek, J.J, Sussman, D, Monnat, R.J, Stoddard, B.L, Baker, D. | | Deposit date: | 2006-01-05 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational redesign of endonuclease DNA binding and cleavage specificity.
Nature, 441, 2006
|
|
6UVW
 
 | |
6UWK
 
 | |
6UWJ
 
 | |
6UWH
 
 | |
6UWG
 
 | |
6UW0
 
 | |
7R9F
 
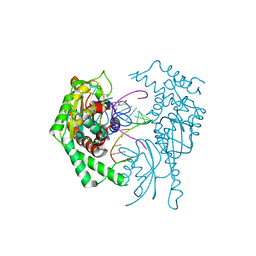 | |
7R9G
 
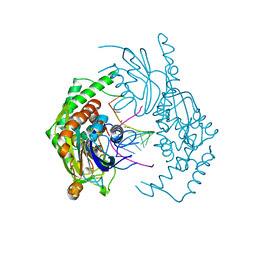 | | Catalytically inactive yeast Pseudouridine Synthase, PUS1, bound to RNA | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*AP*AP*UP*CP*GP*GP*GP*AP*UP*UP*CP*CP*GP*GP*AP*UP*A)-3'), SULFATE ION, ... | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2021-06-29 | | Release date: | 2021-12-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis of mRNA recognition and binding by yeast pseudouridine synthase PUS1.
Plos One, 18, 2023
|
|
1IPP
 
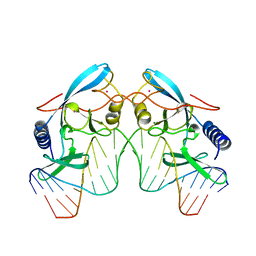 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
7RDR
 
 | |
7RCG
 
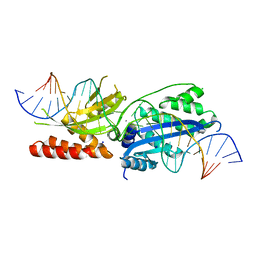 | |
7RCD
 
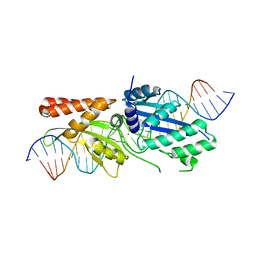 | |
7RCF
 
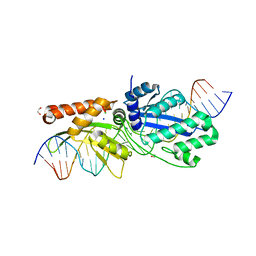 | |
7RCE
 
 | |
7RCC
 
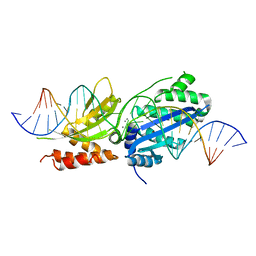 | |
7SQ4
 
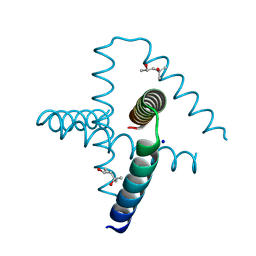 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ3
 
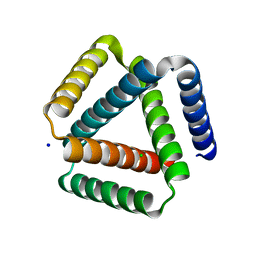 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ5
 
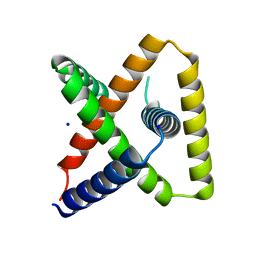 | | Designed trefoil knot protein, variant 3 | | Descriptor: | Designed trefoil knot protein, variant 3, SODIUM ION | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
2O3K
 
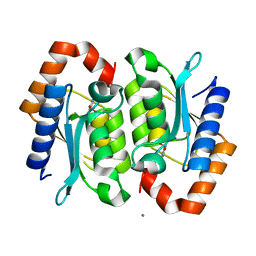 | |
2O6M
 
 | | H98Q mutant of the homing endonuclease I-PPOI complexed with DNA | | Descriptor: | 5'-D(*DTP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DAP*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DA)-3', Intron-encoded endonuclease I-PpoI, MAGNESIUM ION, ... | | Authors: | Eastberg, J.H, Stoddard, B.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism.
Biochemistry, 46, 2007
|
|
7UNH
 
 | |
7UNJ
 
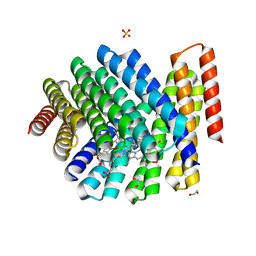 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, SP1-ZnPPaM designed chlorophyll dimer protein, SULFATE ION, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester matching geometry of purple bacterial special pair, SP1-ZnPPaM
To Be Published
|
|
