4AH7
 
 | | Structure of Wild Type Stapylococcus aureus N-acetylneuraminic acid lyase in complex with pyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Timms, N, Poyakova, A, Windle, C.L, Trinh, C.H, Nelson, A, Pearson, A.R, Berry, A. | | Deposit date: | 2012-02-03 | | Release date: | 2013-01-23 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insights Into the Recovery of Aldolase Activity in N-Acetylneuraminic Acid Lyase by Replacement of the Catalytically Active Lysine with Gamma-Thialysine by Using a Chemical Mutagenesis Strategy.
Chembiochem, 14, 2013
|
|
8B37
 
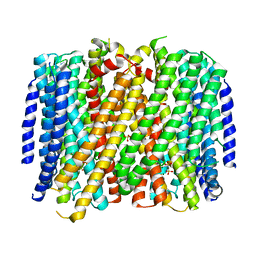 | | Crystal structure of Pyrobaculum aerophilum potassium-independent proton pumping membrane integral pyrophosphatase in complex with imidodiphosphate and magnesium, and with bound sulphate | | Descriptor: | IMIDODIPHOSPHORIC ACID, K(+)-insensitive pyrophosphate-energized proton pump, MAGNESIUM ION, ... | | Authors: | Strauss, J, Wilkinson, C, Vidilaseris, K, Ribeiro, O, Liu, J, Hillier, J, Malinen, A, Gehl, B, Jeuken, L.C, Pearson, A.R, Goldman, A. | | Deposit date: | 2022-09-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Functional and structural asymmetry suggest a unifying principle for catalysis in membrane-bound pyrophosphatases.
Embo Rep., 25, 2024
|
|
5A8G
 
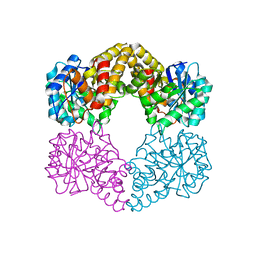 | | Crystal structure of the wild-type Staphylococcus aureus N- acetylneurminic acid lyase in complex with fluoropyruvate | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Stockwell, J, Daniels, A.D, Windle, C.L, Harman, T, Woodhall, T, Trinh, C.H, Lebel, T, Pearson, A.R, Mulholland, K, Berry, A, Nelson, A. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Evaluation of Fluoropyruvate as Nucleophile in Reactions Catalysed by N-Acetyl Neuraminic Acid Lyase Variants: Scope, Limitations and Stereoselectivity.
Org.Biomol.Chem., 14, 2016
|
|
5ZZ1
 
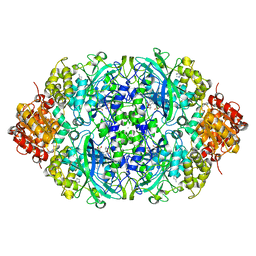 | | Probing the active center of catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, CALCIUM ION, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, ... | | Authors: | Yuzugullu Karakus, Y, Trinh, C.H, Pearson, A.R, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Identification of the site of oxidase substrate binding in Scytalidium thermophilum catalase.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
2WO5
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in space group P21 crystal form I | | Descriptor: | N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WOH
 
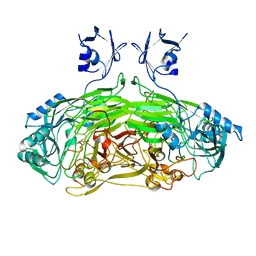 | | Strontium soaked E. coli copper amine oxidase | | Descriptor: | CALCIUM ION, COPPER (II) ION, PRIMARY AMINE OXIDASE, ... | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
2W0Q
 
 | | E. coli copper amine oxidase in complex with Xenon | | Descriptor: | CALCIUM ION, COPPER (II) ION, COPPER AMINE OXIDASE, ... | | Authors: | Pirrat, P, Smith, M.A, Pearson, A.R, McPherson, M.J, Phillips, S.E.V. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structure of a Xenon Derivative of Escherichia Coli Copper Amine Oxidase: Confirmation of the Proposed Oxygen-Entry Pathway.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2WO0
 
 | | EDTA treated E. coli copper amine oxidase | | Descriptor: | COPPER (II) ION, PRIMARY AMINE OXIDASE, SODIUM ION | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-21 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
2WOF
 
 | | EDTA treated E. coli copper amine oxidase | | Descriptor: | COPPER (II) ION, PRIMARY AMINE OXIDASE, SODIUM ION | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
2WKJ
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate at 1.45A resolution in space group P212121 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, PYRUVIC ACID | | Authors: | Campeotto, I, Carr, S.B, Trinh, C.H, Nelson, A.S, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2009-06-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of an Escherichia coli N-acetyl-D-neuraminic acid lyase mutant, E192N, in complex with pyruvate at 1.45 angstrom resolution.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 65, 2009
|
|
2WNZ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 crystal form I | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 2-ETHOXYETHANOL, LACTIC ACID, ... | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNQ
 
 | | Structure of the E192N mutant of E. coli N-acetylneuraminic acid lyase in space group P21 | | Descriptor: | CHLORIDE ION, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-17 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-07-13 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
2WPB
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate and the inhibitor (2R,3R)-2,3,4- trihydroxy-N,N-dipropylbutanamide in space group P21 crystal form I | | Descriptor: | (2R,3R)-2,3,4-TRIHYDROXY-N,N-DIPROPYLBUTANAMIDE, N-ACETYLNEURAMINATE LYASE | | Authors: | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
6EZZ
 
 | | Crystal structure of Escherichia coli amine oxidase mutant E573Q | | Descriptor: | CALCIUM ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gaule, T.G, Smith, M.A, Tych, K.M, Pirrat, P, Trinh, C.H, Pearson, A.R, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxygen Activation Switch in the Copper Amine Oxidase of Escherichia coli.
Biochemistry, 57, 2018
|
|
6FAX
 
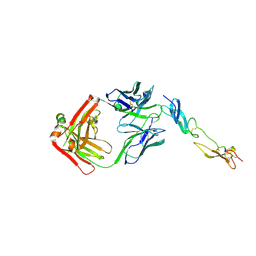 | | Complex of Human CD40 Ectodomain with Lob 7.4 Fab | | Descriptor: | Lob 7.4 heavy chain, Lob 7.4 light chain, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Orr, C.M, Tews, I, Pearson, A.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Complex Interplay between Epitope Specificity and Isotype Dictates the Biological Activity of Anti-human CD40 Antibodies.
Cancer Cell, 33, 2018
|
|
6GRR
 
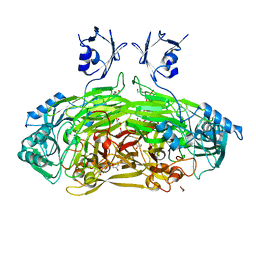 | | Crystal structure of Escherichia coli amine oxidase mutant I342F/E573Q | | Descriptor: | Amine oxidase, CALCIUM ION, COPPER (II) ION, ... | | Authors: | Gaule, T.G, Smith, M.A, Tych, K.M, Pirrat, P, Trinh, C.H, Pearson, A.R, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2018-06-12 | | Release date: | 2018-09-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxygen Activation Switch in the Copper Amine Oxidase of Escherichia coli.
Biochemistry, 57, 2018
|
|
6RXH
 
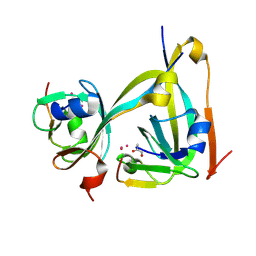 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Aspartate alpha-decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, UNKNOWN ATOM OR ION | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
6RXI
 
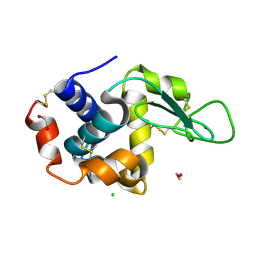 | | In-flow serial synchrotron crystallography using a 3D-printed microfluidic device (3D-MiXD): Lysozyme | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Lysozyme C, ... | | Authors: | Monteiro, D.C.F, von Stetten, D, Pearson, A.R, Trebbin, M. | | Deposit date: | 2019-06-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3D-MiXD: 3D-printed X-ray-compatible microfluidic devices for rapid, low-consumption serial synchrotron crystallography data collection in flow.
Iucrj, 7, 2020
|
|
4C3C
 
 | | Thaumatin refined against hatrx data for time-point 1 | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN-1 | | Authors: | Yorke, B.A, Beddard, G.S, Owen, R.L, Pearson, A.R. | | Deposit date: | 2013-08-22 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Resolved Crystallography Using the Hadamard Transform.
Nat.Methods, 11, 2014
|
|
4BWL
 
 | | Structure of the Y137A mutant of E. coli N-acetylneuraminic acid lyase in complex with pyruvate, N-acetyl-D-mannosamine and N- acetylneuraminic acid | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, 5-(acetylamino)-3,5-dideoxy-D-glycero-D-galacto-non-2-ulosonic acid, N-ACETYLNEURAMINATE LYASE, ... | | Authors: | Campeotto, I, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | Deposit date: | 2013-07-03 | | Release date: | 2014-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Reaction Mechanism of N-Acetylneuraminic Acid Lyase Revealed by a Combination of Crystallography, Qm/Mm Simulation and Mutagenesis.
Acs Chem.Biol., 9, 2014
|
|
4CJ6
 
 | | Crystal structure of the complex of the Cellular Retinal Binding Protein Mutant R234W with 9-cis-retinal | | Descriptor: | RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4CIZ
 
 | | Crystal structure of the complex of the Cellular Retinal Binding Protein with 9-cis-retinal | | Descriptor: | L(+)-TARTARIC ACID, RETINAL, RETINALDEHYDE-BINDING PROTEIN 1 | | Authors: | Bolze, C.S, Helbling, R.E, Owen, R.L, Pearson, A.R, Pompidor, G, Dworkowski, F, Fuchs, M.R, Furrer, J, Golczak, M, Palczewski, K, Cascella, M, Stocker, A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Human Cellular Retinaldehyde-Binding Protein Has Secondary Thermal 9-Cis-Retinal Isomerase Activity.
J.Am.Chem.Soc., 136, 2014
|
|
4B2Y
 
 | | Probing the active center of catalase-phenol oxidase from Scytalidium thermophilum | | Descriptor: | CALCIUM ION, CATALASE-PHENOL OXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yuzugullu, Y, Trinh, C.H, Pearson, A.R, Ogel, Z.B, McPherson, M.J. | | Deposit date: | 2012-07-19 | | Release date: | 2013-04-10 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Investigating the Active Centre of the Scytalidium Thermophilum Catalase
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4AZD
 
 | | T57V mutant of aspartate decarboxylase | | Descriptor: | ASPARTATE 1-DECARBOXYLASE, MALONATE ION | | Authors: | Webb, M.E, Yorke, B.A, Kershaw, T, Lovelock, S, Lobley, C.M.C, Kilkenny, M.L, Smith, A.G, Blundell, T.L, Pearson, A.R, Abell, C. | | Deposit date: | 2012-06-25 | | Release date: | 2012-07-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Threonine 57 is Required for the Post-Translational Activation of Escherichia Coli Aspartate Alpha-Decarboxylase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
