5WJE
 
 | | Crystal structure of Naa80 bound to a bisubstrate analogue | | Descriptor: | Actin N-terminus peptide, CARBOXYMETHYL COENZYME *A, CG8481, ... | | Authors: | Goris, M, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2017-07-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.765 Å) | | Cite: | Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6UIA
 
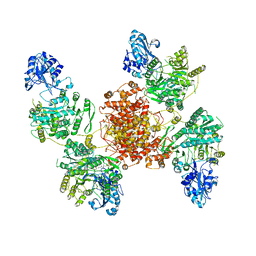 | |
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
1QP9
 
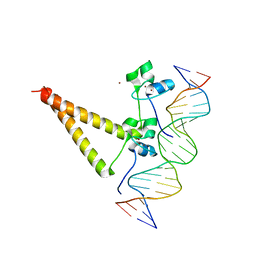 | | STRUCTURE OF HAP1-PC7 COMPLEXED TO THE UAS OF CYC7 | | Descriptor: | CYP1(HAP1-PC7) ACTIVATORY PROTEIN, DNA (5'-D(*AP*CP*GP*CP*TP*AP*TP*TP*AP*TP*CP*GP*CP*TP*AP*TP*TP*AP*GP*T)-3'), DNA (5'-D(*AP*CP*TP*AP*AP*TP*AP*GP*CP*GP*AP*TP*AP*AP*TP*AP*GP*CP*GP*T)-3'), ... | | Authors: | Lukens, A, King, D, Marmorstein, R. | | Deposit date: | 1999-06-01 | | Release date: | 2000-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HAP1-PC7 bound to DNA: implications for DNA recognition and allosteric effects of DNA-binding on transcriptional activation.
Nucleic Acids Res., 28, 2000
|
|
2QIY
 
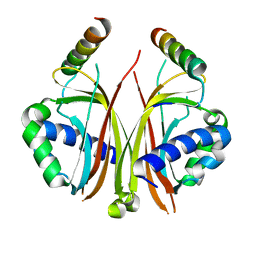 | |
7LLA
 
 | |
7LIW
 
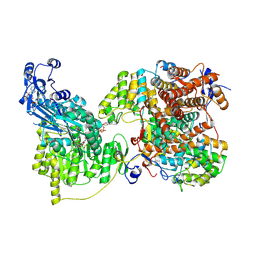 | | Local refinement of human ATP citrate lyase E599Q mutant ASH domain | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2021-01-28 | | Release date: | 2021-05-19 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Reply to: Acetyl-CoA is produced by the citrate synthase homology module of ATP-citrate lyase.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7TOF
 
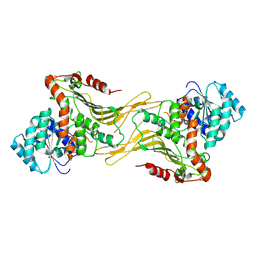 | |
7UAL
 
 | |
7TOE
 
 | |
7UC2
 
 | |
7UAG
 
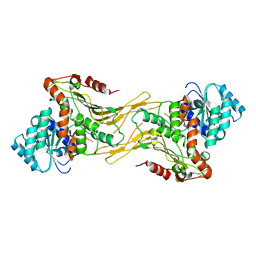 | |
1YGH
 
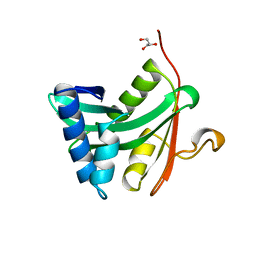 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5ICV
 
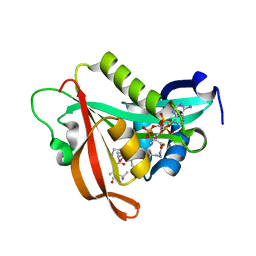 | | Crystal structure of human NatF (hNaa60) bound to a bisubstrate analogue | | Descriptor: | MET-LYS-ALA-VAL-LIG, N-alpha-acetyltransferase 60, [5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)furan-2-yl]methyl (3R)-4-{[3-({(E)-2-[(2,2-dihydroxyethyl)sulfanyl]ethenyl}amino)-3-oxopropyl]amino}-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
5ICW
 
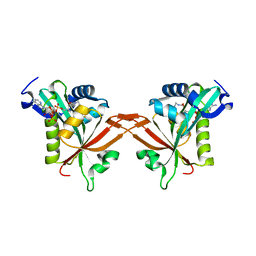 | | Crystal structure of human NatF (hNaa60) homodimer bound to Coenzyme A | | Descriptor: | CHLORIDE ION, COENZYME A, N-alpha-acetyltransferase 60 | | Authors: | Stove, S.I, Magin, R.S, Marmorstein, R, Arnesen, T. | | Deposit date: | 2016-02-23 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of the Golgi-Associated Human N alpha-Acetyltransferase 60 Reveals the Molecular Determinants for Substrate-Specific Acetylation.
Structure, 24, 2016
|
|
5J8C
 
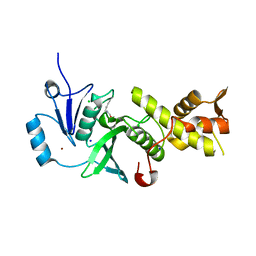 | | Human MOF C316S, E350Q crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, SODIUM ION, ... | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
5J8F
 
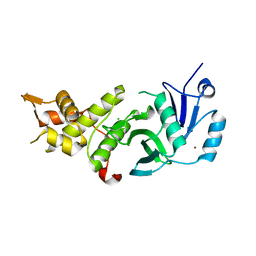 | | Human MOF K274P crystal structure | | Descriptor: | CHLORIDE ION, Histone acetyltransferase KAT8, ZINC ION | | Authors: | McCullough, C.E, Marmorstein, R. | | Deposit date: | 2016-04-07 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Functional Role of Acetyltransferase hMOF K274 Autoacetylation.
J.Biol.Chem., 291, 2016
|
|
1ZX2
 
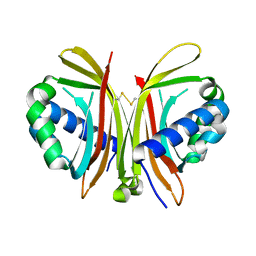 | | Crystal Structure of Yeast UBP3-associated Protein BRE5 | | Descriptor: | UBP3-associated protein BRE5 | | Authors: | Li, K, Zhao, K, Ossareh-Nazari, B, Da, G, Dargemont, C, Marmorstein, R. | | Deposit date: | 2005-06-06 | | Release date: | 2005-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for interaction between the Ubp3 deubiquitinating enzyme and its Bre5 cofactor
J.Biol.Chem., 280, 2005
|
|
5JRQ
 
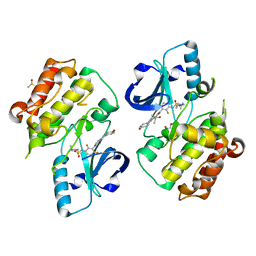 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-6-VEM | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, N-{2,4-difluoro-3-[5-(4-methoxyphenyl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]phenyl}propane-1-sulfonamide, ... | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
5JSM
 
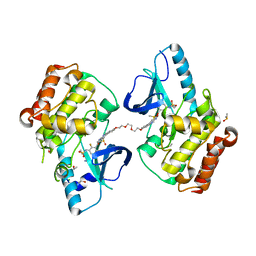 | |
5JT2
 
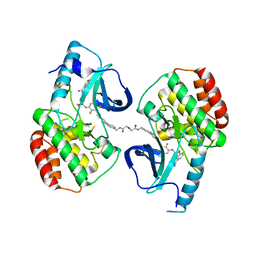 | | BRAFV600E Kinase Domain In Complex with Chemically Linked Vemurafenib Inhibitor VEM-BISAMIDE | | Descriptor: | 2,2'-oxybis(N-{[4-(3-{2,6-difluoro-3-[(propane-1-sulfonyl)amino]benzoyl}-1H-pyrrolo[2,3-b]pyridin-5-yl)phenyl]methyl}acetamide), BENZAMIDINE, Serine/threonine-protein kinase B-raf | | Authors: | Grasso, M.J, Marmorstein, R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Chemically Linked Vemurafenib Inhibitors Promote an Inactive BRAF(V600E) Conformation.
Acs Chem.Biol., 11, 2016
|
|
7KD7
 
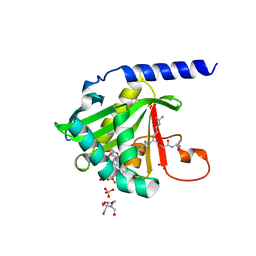 | |
6POF
 
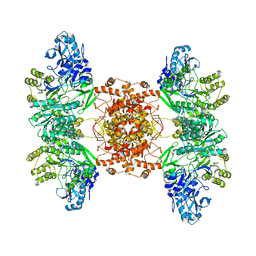 | | Structure of human ATP citrate lyase | | Descriptor: | ATP-citrate synthase | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-07-03 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7KPU
 
 | |
6PPL
 
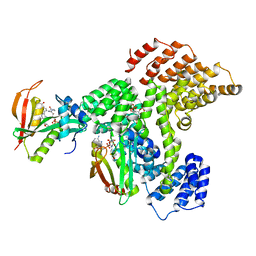 | | Cryo-EM structure of human NatE complex (NatA/Naa50) | | Descriptor: | ACETYL COENZYME *A, INOSITOL HEXAKISPHOSPHATE, N-alpha-acetyltransferase 10, ... | | Authors: | Deng, S, Marmorstein, R. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-19 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Molecular basis for N-terminal acetylation by human NatE and its modulation by HYPK.
Nat Commun, 11, 2020
|
|
