2Q3I
 
 | | Crystal structure of the D10-P3/IQN17 complex: a D-peptide inhibitor of HIV-1 entry bound to the GP41 coiled-coil pocket | | Descriptor: | CHLORIDE ION, D-peptide, Fusion protein between the Coiled-Coil pocket of HIV GP41 and gcn4-PIQI | | Authors: | Malashkevich, V.N, Eckert, D.M, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV Entry: Discovery of D-Peptide Inhibitors that Target the Gp41 Coiled-Coil Pocket
Cell(Cambridge,Mass.), 99, 1999
|
|
3FON
 
 | | Crystal structure of the Class I MHC Molecule H-2Kwm7 with a Single Self Peptide VNDIFEAI | | Descriptor: | Beta-2-microglobulin, MHC, Peptide | | Authors: | Malashkevich, V.N, Qian, J, Jarchum, I, Yamada, T, Mikesh, L, Palmieri, E, Lund, T, Hattori, M, Shabanowitz, J, Hunt, D.F, Ramagopal, U.A, Brims, D.R, Almo, S.C, Nathenson, S.G, DiLorenzo, T.P. | | Deposit date: | 2008-12-30 | | Release date: | 2010-01-12 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect.
Int.Immunol., 22, 2010
|
|
4G2K
 
 | | Crystal structure of the Marburg Virus GP2 ectodomain in its post-fusion conformation | | Descriptor: | CHLORIDE ION, GLYCEROL, General control protein GCN4, ... | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Harrison, J.S, Toro, R, Bhosle, R.C, Chandran, K, Lai, J.R, Almo, S.C. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Marburg Virus GP2 Core Domain in Its Postfusion Conformation.
Biochemistry, 51, 2012
|
|
4KGE
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, pH 4.5 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
4KGF
 
 | | Crystal structure of near-infrared fluorescent protein with an extended stokes shift, ph 8.0 | | Descriptor: | CHLORIDE ION, TagRFP675, red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-29 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Extended Stokes shift in fluorescent proteins: chromophore-protein interactions in a near-infrared TagRFP675 variant.
Sci Rep, 3, 2013
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2Q6Q
 
 | | Crystal structure of Spc42p, a critical component of spindle pole body in budding yeast | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Spindle pole body component SPC42 | | Authors: | Malashkevich, V.N, Newman, J.R, Kim, P.S. | | Deposit date: | 2007-06-05 | | Release date: | 2008-05-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Analysis of coiled-coil interactions between core proteins of the spindle pole body.
Biochemistry, 47, 2008
|
|
3KCS
 
 | | Crystal structure of PAmCherry1 in the dark state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3KCT
 
 | | CRYSTAL STRUCTURE OF PAmCherry1 in the photoactivated state | | Descriptor: | PAmCherry1 protein | | Authors: | Malashkevich, V.N, Subach, F.V, Zencheck, W.D, Xiao, H, Filonov, G.S, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Photoactivation mechanism of PAmCherry based on crystal structures of the protein in the dark and fluorescent states.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3M22
 
 | | Crystal structure of TagRFP fluorescent protein | | Descriptor: | TagRFP | | Authors: | Malashkevich, V.N, Subach, O.M, Ramagopal, U.A, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2010-03-06 | | Release date: | 2010-05-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins.
Chem.Biol., 17, 2010
|
|
3M24
 
 | | Crystal structure of TagBFP fluorescent protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Malashkevich, V.N, Subach, O.M, Almo, S.C, Verkhusha, V.V. | | Deposit date: | 2010-03-06 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins.
Chem.Biol., 17, 2010
|
|
3NT9
 
 | |
3NT3
 
 | | CRYSTAL STRUCTURE OF LSSmKate2 red fluorescent proteins with large Stokes shift | | Descriptor: | GLYCEROL, LSSmKate2 red fluorescent protein | | Authors: | Malashkevich, V.N, Piatkevich, K, Almo, S.C, Verkhusha, V. | | Deposit date: | 2010-07-02 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering ESPT Pathways Based on Structural Analysis of LSSmKate Red Fluorescent Proteins with Large Stokes Shift.
J.Am.Chem.Soc., 132, 2010
|
|
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | Descriptor: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDS
 
 | | Crystal structure of the refolded R7-2 streptavidin | | Descriptor: | PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDQ
 
 | | Crystal structure of R7-2 streptavidin complexed with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4N21
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
4XTQ
 
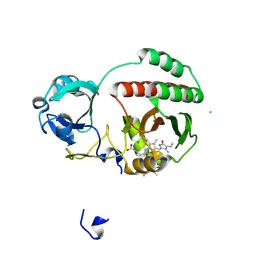 | | Crystal structure of a mutant (C20S) of a near-infrared fluorescent protein BphP1-FP | | Descriptor: | 3-[2-[(Z)-[5-[(Z)-[(3R,4R)-3-ethenyl-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-pyrrol-2-ylidene]methyl]-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, BphP1-FP/C20S, CHLORIDE ION | | Authors: | Pletnev, S, Malashkevich, V.N. | | Deposit date: | 2015-01-23 | | Release date: | 2015-12-09 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins.
Chem.Biol., 22, 2015
|
|
1PIQ
 
 | | CRYSTAL STRUCTURE OF GCN4-PIQ, A TRIMERIC COILED COIL WITH BURIED POLAR RESIDUES | | Descriptor: | CHLORIDE ION, PROTEIN (GENERAL CONTROL PROTEIN GCN4-PIQ) | | Authors: | Eckert, D.M, Malashkevich, V.N, Kim, P.S. | | Deposit date: | 1998-09-25 | | Release date: | 1998-09-30 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of GCN4-pIQI, a trimeric coiled coil with buried polar residues.
J.Mol.Biol., 284, 1998
|
|
1FBM
 
 | | ASSEMBLY DOMAIN OF CARTILAGE OLIGOMERIC MATRIX PROTEIN IN COMPLEX WITH ALL-TRANS RETINOL | | Descriptor: | PROTEIN (CARTILAGE OLIGOMERIC MATRIX PROTEIN), RETINOL | | Authors: | Guo, Y, Bozic, D, Malashkevich, V.N, Kammerer, R.A, Schulthess, T. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | All-trans retinol, vitamin D and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein.
EMBO J., 17, 1998
|
|
1JS3
 
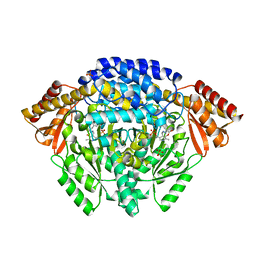 | | Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa | | Descriptor: | CARBIDOPA, DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
1JS6
 
 | | Crystal Structure of DOPA decarboxylase | | Descriptor: | DOPA decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Burkhard, P, Dominici, P, Borri-Voltattorni, C, Jansonius, J.N, Malashkevich, V.N. | | Deposit date: | 2001-08-16 | | Release date: | 2001-10-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase.
Nat.Struct.Biol., 8, 2001
|
|
1CZQ
 
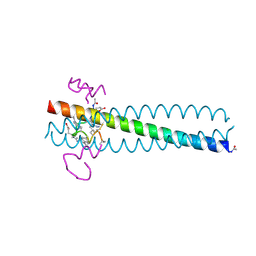 | | CRYSTAL STRUCTURE OF THE D10-P1/IQN17 COMPLEX: A D-PEPTIDE INHIBITOR OF HIV-1 ENTRY BOUND TO THE GP41 COILED-COIL POCKET. | | Descriptor: | CHLORIDE ION, D-PEPTIDE INHIBITOR, FUSION PROTEIN BETWEEN THE HYDROPHOBIC POCKET OF HIV GP41 AND GCN4-PIQI | | Authors: | Eckert, D.M, Malashkevich, V.N, Hong, L.H, Carr, P.A, Kim, P.S. | | Deposit date: | 1999-09-06 | | Release date: | 1999-10-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket.
Cell(Cambridge,Mass.), 99, 1999
|
|
