4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4REF
 
 | | Crystal Structure of TR3 LBD_L449W in complex with Molecule 2 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)hexan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
4RZE
 
 | | Crystal Structure Analysis of the NUR77 Ligand Binding Domain, L437W,D594E mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Fengwei, L, Xuyang, T, Anzhong, L, Li, L, Yuan, L, Hangzi, C, Qiao, W, Tianwei, L. | | Deposit date: | 2014-12-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Impeding the interaction between Nur77 and p38 reduces LPS-induced inflammation.
Nat.Chem.Biol., 11, 2015
|
|
8ARD
 
 | |
5I0I
 
 | | Crystal structure of myosin X motor domain with 2IQ motifs in pre-powerstroke state | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Calmodulin, ... | | Authors: | Isabet, T, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-09-07 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
5I0H
 
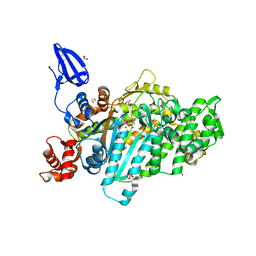 | | Crystal structure of myosin X motor domain in pre-powerstroke state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Isabet, T, Blanc, F, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-02-04 | | Release date: | 2016-09-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
4NNG
 
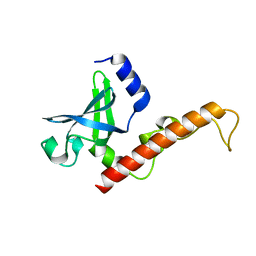 | |
4NNH
 
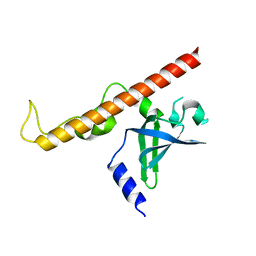 | |
4NNI
 
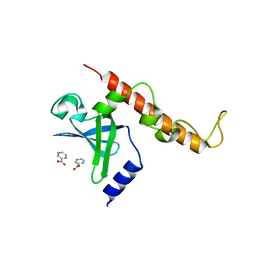 | | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide | | Descriptor: | 30S ribosomal protein S1, PYRAZINE-2-CARBOXYLIC ACID | | Authors: | Yang, J, Liu, Y, Cai, Q, Lin, D. | | Deposit date: | 2013-11-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for targeting the ribosomal protein S1 of Mycobacterium tuberculosis by pyrazinamide.
Mol.Microbiol., 95, 2015
|
|
4NNK
 
 | |
6LKA
 
 | | Crystal Structure of EV71-3C protease with a Novel Macrocyclic Compounds | | Descriptor: | 3C proteinase, ~{N}-[(2~{S})-1-[[(2~{S},3~{S},6~{S},7~{Z},12~{E})-4,9-bis(oxidanylidene)-6-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]-2-phenyl-1,10-dioxa-5-azacyclopentadeca-7,12-dien-3-yl]amino]-3-methyl-1-oxidanylidene-butan-2-yl]-5-methyl-1,2-oxazole-3-carboxamide | | Authors: | Li, P, Wu, S.Q, Xiao, T.Y.C, Li, Y.L, Su, Z.M, Hao, F, Hu, G.P, Hu, J, Lin, F.S, Chen, X.S, Gu, Z.X, He, H.Y, Li, J, Chen, S.H. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Design, synthesis, and evaluation of a novel macrocyclic anti-EV71 agent.
Bioorg.Med.Chem., 28, 2020
|
|
5HMO
 
 | |
5HMP
 
 | | Myosin Vc pre-powerstroke state | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, DIMETHYL SULFOXIDE, ... | | Authors: | Ropars, V, Pylypenko, O, Sweeney, H.L, Houdusse, A. | | Deposit date: | 2016-01-16 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | The myosin X motor is optimized for movement on actin bundles.
Nat Commun, 7, 2016
|
|
5KG8
 
 | |
2OO9
 
 | |
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4DZ6
 
 | |
4DI6
 
 | |
3QMX
 
 | |
5TOZ
 
 | | JAK3 with covalent inhibitor PF-06651600 | | Descriptor: | 1-{(2S,5R)-2-methyl-5-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a JAK3-Selective Inhibitor: Functional Differentiation of JAK3-Selective Inhibition over pan-JAK or JAK1-Selective Inhibition.
ACS Chem. Biol., 11, 2016
|
|
5TTU
 
 | | Jak3 with covalent inhibitor 7 | | Descriptor: | 1-[(3aR,7aR)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)octahydro-6H-pyrrolo[2,3-c]pyridin-6-yl]propan-1-one, SULFATE ION, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
5TTS
 
 | | Jak3 with covalent inhibitor 4 | | Descriptor: | 1-{(3R)-3-[(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}propan-1-one, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
5TTV
 
 | | Jak3 with covalent inhibitor 6 | | Descriptor: | N-[3-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)phenyl]propanamide, Tyrosine-protein kinase JAK3 | | Authors: | Vajdos, F.F. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of a Janus Kinase 3 (JAK3) Specific Inhibitor 1-((2S,5R)-5-((7H-Pyrrolo[2,3-d]pyrimidin-4-yl)amino)-2-methylpiperidin-1-yl)prop-2-en-1-one (PF-06651600) Allowing for the Interrogation of JAK3 Signaling in Humans.
J. Med. Chem., 60, 2017
|
|
4L79
 
 | | Crystal Structure of nucleotide-free Myosin 1b residues 1-728 with bound Calmodulin | | Descriptor: | Calmodulin, MAGNESIUM ION, Unconventional myosin-Ib | | Authors: | Shuman, H, Zwolak, A, Dominguez, R, Ostap, E.M. | | Deposit date: | 2013-06-13 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A vertebrate myosin-I structure reveals unique insights into myosin mechanochemical tuning.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OOA
 
 | |
