6AAX
 
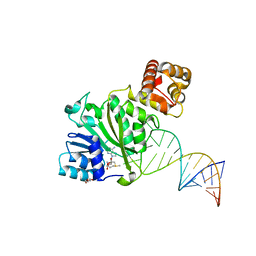 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6AJK
 
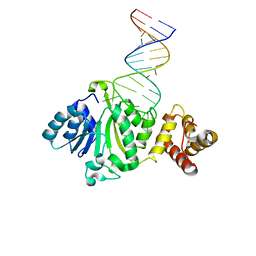 | | Crystal structure of TFB1M and h45 in homo sapiens | | Descriptor: | Dimethyladenosine transferase 1, mitochondrial, RNA (28-MER) | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Wang, C, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
6B7N
 
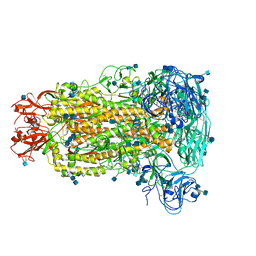 | | Cryo-electron microscopy structure of porcine delta coronavirus spike protein in the pre-fusion state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shang, J, Zheng, Y, Yang, Y, Liu, C, Geng, Q, Tai, W, Du, L, Zhou, Y, Zhang, W, Li, F. | | Deposit date: | 2017-10-04 | | Release date: | 2017-10-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy Structure of Porcine Deltacoronavirus Spike Protein in the Prefusion State
J. Virol., 92, 2018
|
|
5VST
 
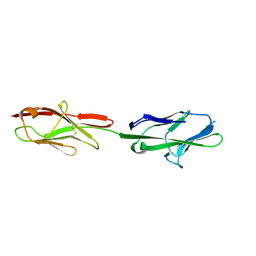 | | Crystal structure of murine CEACAM1b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Biliary glycoprotein | | Authors: | Peng, G, Yang, Y, Pasquarella, J.R, Xu, L, Qian, Z, Holmes, K.V, Li, F. | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism for coronavirus-driven evolution of mouse receptor
J. Biol. Chem., 292, 2017
|
|
6BV2
 
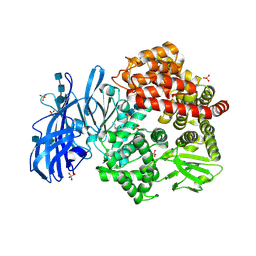 | | Crystal structure of porcine aminopeptidase-N with Isoleucine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV4
 
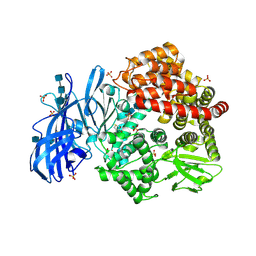 | | Crystal structure of porcine aminopeptidase-N with Methionine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BUY
 
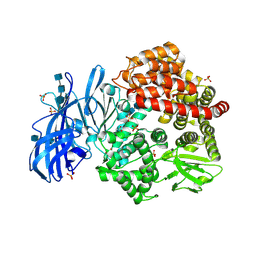 | | Crystal structure of porcine aminopeptidase-N with Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-11 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV0
 
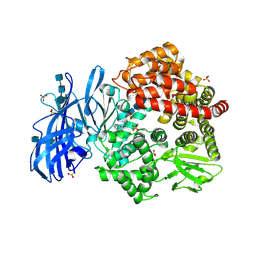 | | Crystal structure of porcine aminopeptidase-N with Arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
6BV1
 
 | | Crystal structure of porcine aminopeptidase-N with Aspartic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L, Lin, Y.-L, Li, F. | | Deposit date: | 2017-12-12 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Rational Design of Therapeutic Peptides for Aminopeptidase N using a Substrate-Based Approach.
Sci Rep, 7, 2017
|
|
7VCT
 
 | | Human p97 single hexamer conformer III with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCS
 
 | | Human p97 double hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCX
 
 | | Human p97 single hexamer conformer II with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCU
 
 | | Human p97 double hexamer conformer I with D1-ATPgammaS and D2-ADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VCV
 
 | | Human p97 single hexamer conformer I with ATPgammaS bound | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Gao, H, Li, F, Shi, Z, Li, Y, Yu, H. | | Deposit date: | 2021-09-04 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures of human p97 double hexamer capture potentiated ATPase-competent state.
Cell Discov, 8, 2022
|
|
7VL9
 
 | | Cryo-EM structure of the CCL15(26-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(26-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VLA
 
 | | Cryo-EM structure of the CCL15(27-92) bound CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CCL15(27-92), CHOLESTEROL, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7VL8
 
 | | Cryo-EM structure of the Apo CCR1-Gi complex | | Descriptor: | C-C chemokine receptor type 1, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Shao, Z, Shen, Q, Mao, C, Yao, B, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Ma, H, Chen, Z, Xu, H.E, Ying, S, Zhang, Y, Shen, H. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Identification and mechanism of G protein-biased ligands for chemokine receptor CCR1.
Nat.Chem.Biol., 18, 2022
|
|
7X9Y
 
 | | Cryo-EM structure of the apo CCR3-Gi complex | | Descriptor: | C-C chemokine receptor type 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-16 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7XA3
 
 | | Cryo-EM structure of the CCL2 bound CCR2-Gi complex | | Descriptor: | C-C motif chemokine 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Shao, Z, Tan, Y, Shen, Q, Yao, B, Hou, L, Qin, J, Xu, P, Mao, C, Chen, L, Zhang, H, Shen, D, Zhang, C, Li, W, Du, X, Li, F, Chen, Z, Jiang, Y, Xu, H.E, Ying, S, Ma, H, Zhang, Y, Shen, H. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular insights into ligand recognition and activation of chemokine receptors CCR2 and CCR3.
Cell Discov, 8, 2022
|
|
7EN0
 
 | | Structure and Activity of SLAC1 Channels for Stomatal Signaling in Leaves | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, SLow Anion Channel 1, SPHINGOSINE | | Authors: | Deng, Y, Kashtoh, H, Wang, Q, Zhen, G, Li, Q, Tang, L, Gao, H, Zhang, C, Qin, L, Su, M, Li, F, Huang, X, Wang, Y, Xie, Q, Clarke, O.B, Hendrickson, W.A, Chen, Y. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and activity of SLAC1 channels for stomatal signaling in leaves.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8HKD
 
 | |
8HOJ
 
 | | Crystal structure of UGT71AP2 in complex with UDP | | Descriptor: | UGT71AP2, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | crystal structure of UGT71AP2 in complex with UDP
To Be Published
|
|
8HOK
 
 | | crystal structure of UGT71AP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, UGT71AP2 | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | crystal structure of UGT71AP2
To Be Published
|
|
8J23
 
 | | Cryo-EM structure of FFAR2 complex in apo state | | Descriptor: | Free fatty acid receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Tai, L, Li, F, Sun, X, Tang, W, Wang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular recognition and activation mechanism of short chain fatty acid receptors FFAR2 and FFAR3
To Be Published
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
