6E2M
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-methyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2N
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | Mitogen-activated protein kinase kinase kinase 5, N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl][3,4'-bipyridine]-2'-carboxamide | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6E2O
 
 | | ASK1 kinase domain complex with inhibitor | | Descriptor: | 4-(4-cyclopropyl-1H-imidazol-1-yl)-N-[3-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)phenyl]pyridine-2-carboxamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Lansdon, E.B. | | Deposit date: | 2018-07-11 | | Release date: | 2018-09-19 | | Last modified: | 2018-10-17 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | ASK1 contributes to fibrosis and dysfunction in models of kidney disease.
J. Clin. Invest., 128, 2018
|
|
6KBN
 
 | | Crystal structure of Vac8 (del 19-33) bound to Atg13 | | Descriptor: | Autophagy-related protein 13, Vacuolar protein 8 | | Authors: | Park, J, Lee, C. | | Deposit date: | 2019-06-25 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways.
Autophagy, 16, 2020
|
|
6KV1
 
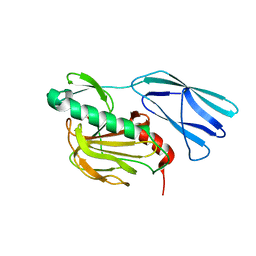 | | Structure of wild type closed form of peptidoglycan peptidase ZN SAD | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-09-03 | | Release date: | 2020-01-15 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.722 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
3KQI
 
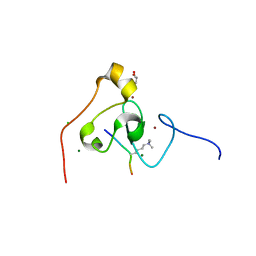 | | crystal structure of PHF2 PHD domain complexed with H3K4Me3 peptide | | Descriptor: | CHLORIDE ION, GLYCEROL, H3K4Me3 peptide, ... | | Authors: | Wen, H, Li, J.Z, Song, T, Lu, M, Lee, M. | | Deposit date: | 2009-11-17 | | Release date: | 2010-02-02 | | Last modified: | 2019-02-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation.
J.Biol.Chem., 285, 2010
|
|
7T7G
 
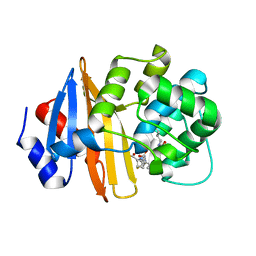 | | Imipenem-OXA-23 2 minute complex | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7E
 
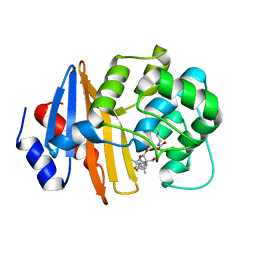 | | MA-1-206-OXA-23 3 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7D
 
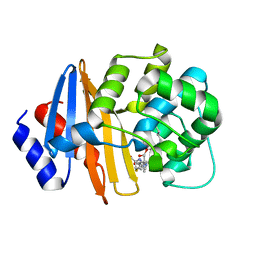 | | MA-1-206-OXA-23 30s complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
7T7F
 
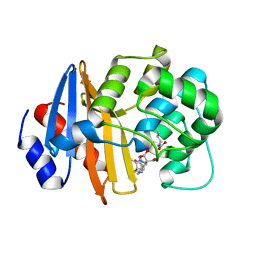 | | MA-1-206-OXA-23 25 minute complex | | Descriptor: | (2R,4S)-2-(1,3-dihydroxypropan-2-yl)-4-{[(3R,5R)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase OXA-23 | | Authors: | Smith, C.A, Stewart, N.K, Vakulenko, S.B. | | Deposit date: | 2021-12-15 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C6 Hydroxymethyl-Substituted Carbapenem MA-1-206 Inhibits the Major Acinetobacter baumannii Carbapenemase OXA-23 by Impeding Deacylation.
Mbio, 13, 2022
|
|
8C54
 
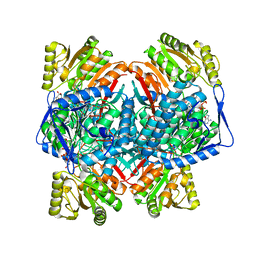 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
8DA2
 
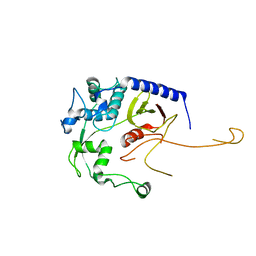 | | Acinetobacter baumannii L,D-transpeptidase | | Descriptor: | L,D-transpeptidase family protein | | Authors: | Toth, M, Stewart, N.K, Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2022-06-12 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The l,d-Transpeptidase Ldt Ab from Acinetobacter baumannii Is Poorly Inhibited by Carbapenems and Has a Unique Structural Architecture.
Acs Infect Dis., 8, 2022
|
|
2JTN
 
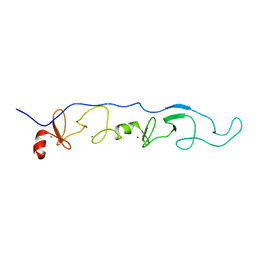 | | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | | Descriptor: | LIM domain-binding protein 1, LIM/homeobox protein Lhx3, ZINC ION | | Authors: | Lee, C, Nancarrow, A.L, Mackay, J.P, Matthews, J.M. | | Deposit date: | 2007-08-03 | | Release date: | 2008-06-17 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes
Embo J., 27, 2008
|
|
6Z1X
 
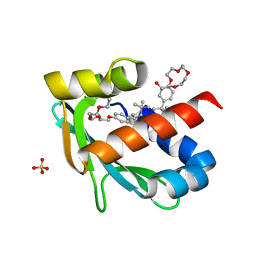 | |
6Z1W
 
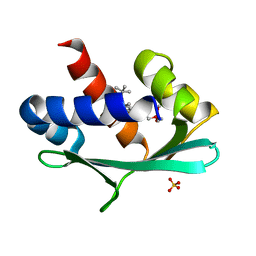 | |
5C22
 
 | | Crystal structure of Zn-bound HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D, ZINC ION | | Authors: | Ha, N.C, Kim, J.S. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
5C21
 
 | | Crystal structure of native HlyD from E. coli | | Descriptor: | Chromosomal hemolysin D | | Authors: | Ha, N.C, Kim, J.S, Yoon, B.Y. | | Deposit date: | 2015-06-15 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Soluble Fragment of the Membrane Fusion Protein HlyD in a Type I Secretion System of Gram-Negative Bacteria
Structure, 24, 2016
|
|
7LRQ
 
 | | Crystal structure of human SFPQ/NONO heterodimer, conserved DBHS region | | Descriptor: | CHLORIDE ION, Non-POU domain-containing octamer-binding protein, Splicing factor, ... | | Authors: | Marshall, A.C, Bond, C.S, Mohnen, I. | | Deposit date: | 2021-02-17 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paraspeckle subnuclear bodies depend on dynamic heterodimerisation of DBHS RNA-binding proteins via their structured domains.
J.Biol.Chem., 298, 2022
|
|
5XJG
 
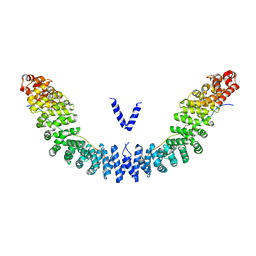 | | Crystal structure of Vac8p bound to Nvj1p | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Nucleus-vacuole junction protein 1, ... | | Authors: | Jeong, H, Park, J, Jun, Y, Lee, C. | | Deposit date: | 2017-05-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8DUA
 
 | | SIV E660.CR54 SOS-2P Env Trimer with ITS92.02 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, Envelope glycoprotein gp41, ... | | Authors: | Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-27 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.32 Å) | | Cite: | Cryo-EM structures of prefusion SIV envelope trimer.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8DVD
 
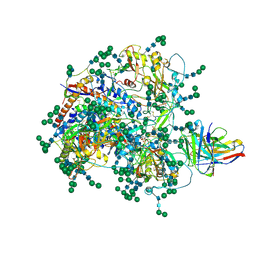 | |
8ELI
 
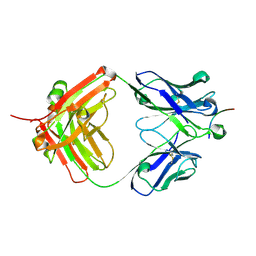 | |
8EUW
 
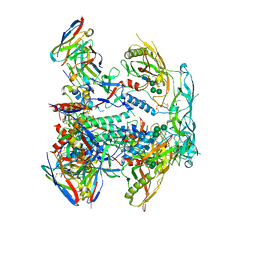 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-MM28 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUU
 
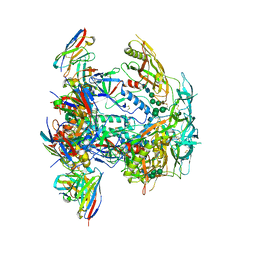 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
8EUV
 
 | | Cryo-EM structure of HIV-1 BG505 DS-SOSIP ENV trimer bound to VRC34.01-COMBO1 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Pletnev, S, Kwong, P. | | Deposit date: | 2022-10-19 | | Release date: | 2023-09-27 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Antibody-directed evolution reveals a mechanism for enhanced neutralization at the HIV-1 fusion peptide site.
Nat Commun, 14, 2023
|
|
