5WDR
 
 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
5WDP
 
 | | H-Ras mutant L120A bound to GMP-PNP at 277K | | Descriptor: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Cofsky, J.C, Bandaru, P, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
1PLR
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
1PLQ
 
 | | CRYSTAL STRUCTURE OF THE EUKARYOTIC DNA POLYMERASE PROCESSIVITY FACTOR PCNA | | Descriptor: | MERCURY (II) ION, PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) | | Authors: | Krishna, T.S.R, Kong, X.-P, Gary, S, Burgers, P.M, Kuriyan, J. | | Deposit date: | 1995-01-02 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA.
Cell(Cambridge,Mass.), 79, 1994
|
|
3LAH
 
 | | Structural insights into the molecular mechanism of H-NOX activation | | Descriptor: | IMIDAZOLE, Methyl-accepting chemotaxis protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Olea Jr, C, Herzik Jr, M.A, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the molecular mechanism of H-NOX activation.
Protein Sci., 19, 2010
|
|
3NVR
 
 | | Modulating Heme Redox Potential Through Protein-Induced Porphyrin Distortion | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Olea Jr, C, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Modulating heme redox potential through protein-induced porphyrin distortion
J.Am.Chem.Soc., 132, 2010
|
|
1EFN
 
 | |
3NVU
 
 | | Modulating Heme Redox Potential Through Protein-Induced Porphyrin Distortion | | Descriptor: | CHLORIDE ION, Methyl-accepting chemotaxis protein, OXYGEN MOLECULE, ... | | Authors: | Olea Jr, C, Kuriyan, J, Marletta, M.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | Modulating heme redox potential through protein-induced porphyrin distortion.
J.Am.Chem.Soc., 132, 2010
|
|
3ET6
 
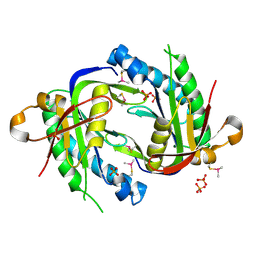 | | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase | | Descriptor: | PHOSPHATE ION, Soluble guanylyl cyclase beta | | Authors: | Winger, J.A, Derbyshire, E.R, Lamers, M.H, Marletta, M.A, Kuriyan, J. | | Deposit date: | 2008-10-07 | | Release date: | 2008-10-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Bmc Struct.Biol., 8, 2008
|
|
2RFD
 
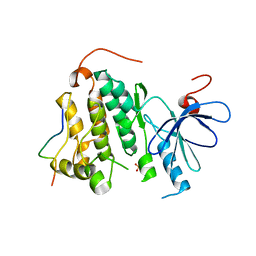 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor, SULFATE ION | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RF9
 
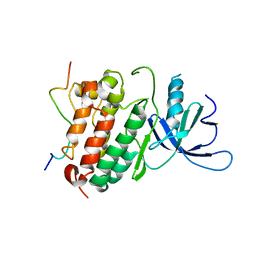 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
2RFE
 
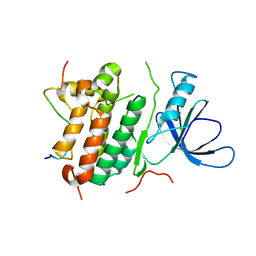 | | Crystal structure of the complex between the EGFR kinase domain and a Mig6 peptide | | Descriptor: | ERBB receptor feedback inhibitor 1, Epidermal growth factor receptor | | Authors: | Zhang, X, Pickin, K.A, Bose, R, Jura, N, Cole, P.A, Kuriyan, J. | | Deposit date: | 2007-09-28 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface.
Nature, 450, 2007
|
|
1TDE
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THIOREDOXIN REDUCTASE REFINED AT 2 ANGSTROM RESOLUTION: IMPLICATIONS FOR A LARGE CONFORMATIONAL CHANGE DURING CATALYSIS | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Waksman, G, Krishna, T.S.R, Williams Junior, C.H, Kuriyan, J. | | Deposit date: | 1994-01-14 | | Release date: | 1994-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli thioredoxin reductase refined at 2 A resolution. Implications for a large conformational change during catalysis.
J.Mol.Biol., 236, 1994
|
|
3SOA
 
 | | Full-length human CaMKII | | Descriptor: | 4-[(2,4-dichloro-5-methoxyphenyl)amino]-6-methoxy-7-[3-(4-methylpiperazin-1-yl)propoxy]quinoline-3-carbonitrile, Calcium/calmodulin-dependent protein kinase type II subunit alpha with a beta 7 linker | | Authors: | Chao, L.H, Kuriyan, J. | | Deposit date: | 2011-06-30 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5501 Å) | | Cite: | A Mechanism for Tunable Autoinhibition in the Structure of a Human Ca(2+)/Calmodulin- Dependent Kinase II Holoenzyme.
Cell(Cambridge,Mass.), 146, 2011
|
|
3GEQ
 
 | | Structural basis for the chemical rescue of Src kinase activity | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Muratore, K.E, Seeliger, M.A, Wang, Z, Fomina, D, Neiswinger, J, Havranek, J.J, Baker, D, Kuriyan, J, Cole, P.A. | | Deposit date: | 2009-02-25 | | Release date: | 2009-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparative analysis of mutant tyrosine kinase chemical rescue.
Biochemistry, 48, 2009
|
|
3KSY
 
 | |
2GS6
 
 | | Crystal Structure of the active EGFR kinase domain in complex with an ATP analog-peptide conjugate | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, Peptide, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An Allosteric Mechanism for Activation of the Kinase Domain of Epidermal Growth Factor Receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS7
 
 | | Crystal Structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor
Cell(Cambridge,Mass.), 125, 2006
|
|
2GS2
 
 | | Crystal Structure of the active EGFR kinase domain | | Descriptor: | Epidermal growth factor receptor | | Authors: | Zhang, X, Gureasko, J, Shen, K, Cole, P.A, Kuriyan, J. | | Deposit date: | 2006-04-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor.
Cell(Cambridge,Mass.), 125, 2006
|
|
2OIQ
 
 | | Crystal Structure of chicken c-Src kinase domain in complex with the cancer drug imatinib. | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Seeliger, M.A, Nagar, B, Frank, F, Cao, X, Henderson, M.N, Kuriyan, J. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | c-Src Binds to the Cancer Drug Imatinib with an Inactive Abl/c-Kit Conformation and a Distributed Thermodynamic Penalty.
Structure, 15, 2007
|
|
2OZO
 
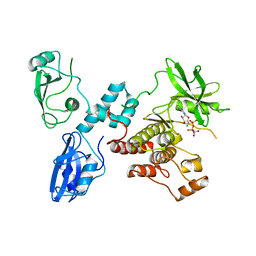 | | Autoinhibited intact human ZAP-70 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase ZAP-70 | | Authors: | Deindl, S, Kadlecek, T.A, Brdicka, T, Cao, X, Weiss, A, Kuriyan, J. | | Deposit date: | 2007-02-26 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for the Inhibition of Tyrosine Kinase Activity of ZAP-70.
Cell(Cambridge,Mass.), 129, 2007
|
|
2POL
 
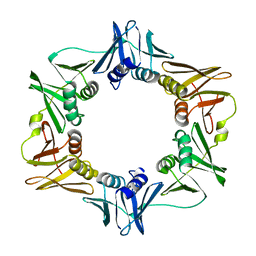 | |
2F86
 
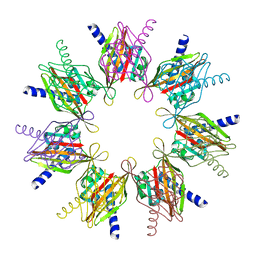 | |
1IAS
 
 | | CYTOPLASMIC DOMAIN OF UNPHOSPHORYLATED TYPE I TGF-BETA RECEPTOR CRYSTALLIZED WITHOUT FKBP12 | | Descriptor: | SULFATE ION, TGF-BETA RECEPTOR TYPE I | | Authors: | Huse, M, Muir, T.W, Chen, Y.-G, Kuriyan, J, Massague, J. | | Deposit date: | 2001-03-23 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The TGF beta receptor activation process: an inhibitor- to substrate-binding switch.
Mol.Cell, 8, 2001
|
|
3BEP
 
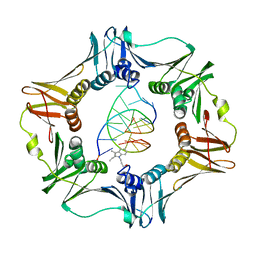 | | Structure of a sliding clamp on DNA | | Descriptor: | 1-(3-hydroxypropyl)-2-{(1E,3E,5E)-5-[1-(3-hydroxypropyl)-3,3-dimethyl-1,3-dihydro-2H-indol-2-ylidene]penta-1,3-dien-1-y l}-3,3-dimethyl-3H-indolium, DNA (5'-D(*DTP*DTP*DTP*DTP*DAP*DTP*DAP*DCP*DGP*DAP*DTP*DGP*DGP*DG)-3'), DNA (5'-D(P*DCP*DCP*DCP*DAP*DTP*DCP*DGP*DTP*DAP*DT)-3'), ... | | Authors: | Georgescu, R.E, Kim, S.S, Yurieva, O, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2007-11-19 | | Release date: | 2008-01-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of a sliding clamp on DNA
Cell(Cambridge,Mass.), 132, 2008
|
|
