2NYX
 
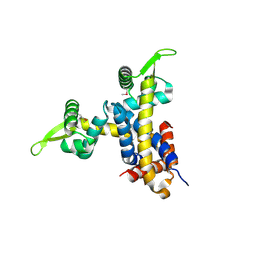 | | Crystal structure of RV1404 from Mycobacterium tuberculosis | | Descriptor: | Probable transcriptional regulatory protein, Rv1404 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, R, Kim, C.-Y, Kaviratne, T, Woodruff, T, Segelke, B.W, Lekin, T, Toppani, D, Terwilliger, T.C, Hung, L.-W, TB Structural Genomics Consortium (TBSGC), Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of RV1404 from Mycobacterium tuberculosis
To be Published
|
|
2G2D
 
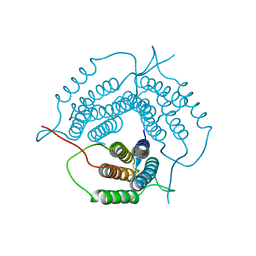 | | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis | | Descriptor: | ATP:cobalamin adenosyltransferase | | Authors: | Moon, J.H, Kaviratne, A, Yu, M, Bursey, E.H, Hung, L.-W, Lekin, T.P, Segelke, B.W, Terwilliger, T.C, Kim, C.-Y, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-15 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative pduO-type ATP:cobalamin adenosyltransferase from Mycobacterium tuberculosis.
To be Published
|
|
2A2J
 
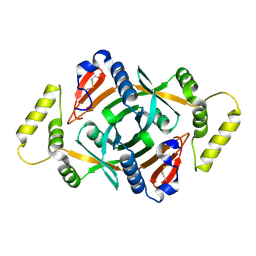 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
3RGA
 
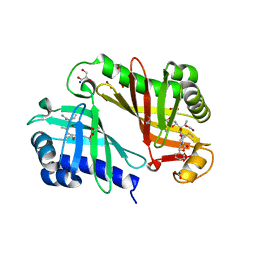 | | Crystal structure of epoxide hydrolase for polyether lasalocid A biosynthesis | | Descriptor: | (4R,5S)-3-[(2R)-2-{(2S,2'R,4S,5S,5'R)-2,5'-diethyl-5'-[(1S)-1-hydroxyethyl]-4-methyloctahydro-2,2'-bifuran-5-yl}butanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, (4R,5S)-3-[(2R,3S,4S)-2-ethyl-5-[(3R)-2-ethyl-3-[2-[(2R,3R)-2-ethyl-3-methyl-oxiran-2-yl]ethyl]oxiran-2-yl]-3-hydroxy-4-methyl-pentanoyl]-4-methyl-5-phenyl-1,3-oxazolidin-2-one, ACETATE ION, ... | | Authors: | Hotta, K, Mathews, I.I, Chen, X, Kim, C.-Y. | | Deposit date: | 2011-04-08 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enzymatic catalysis of anti-Baldwin ring closure in polyether biosynthesis
Nature, 483, 2012
|
|
6O7F
 
 | |
3B4W
 
 | | Crystal structure of Mycobacterium tuberculosis aldehyde dehydrogenase complexed with NAD+ | | Descriptor: | Aldehyde dehydrogenase, ETHANOL, GLYCEROL, ... | | Authors: | Moon, J.H, Lyon, A.E, Yu, M, Hung, L.-W, Terwilliger, T, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-10-24 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of aldehyde dehydrogenase from Mycobacterium tuberculosis complexed with NAD+.
To Be Published
|
|
3LP6
 
 | | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A resolution | | Descriptor: | FORMIC ACID, GLYCEROL, Phosphoribosylaminoimidazole carboxylase catalytic subunit | | Authors: | Kim, H, Yu, M, Hung, L.-W, Terwilliger, T.C, Kim, C.-Y, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Crystal Structure of Rv3275c-E60A from Mycobacterium tuberculosis at 1.7A
Resolution
To be Published
|
|
1W30
 
 | | PyrR of Mycobacterium Tuberculosis as a potential drug target | | Descriptor: | PYRR BIFUNCTIONAL PROTEIN | | Authors: | Kantardjieff, K.A, Vasquez, C, Castro, P, Warfel, N.N, Rho, B.-S, Lekin, T, Kim, C.-Y, Segelke, B.W, Terwilliger, T, Rupp, B, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Pyrr (Rv1379) from Mycobacterium Tuberculosis: A Persistence Gene and Protein Drug Target
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1I9N
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,5-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,5-DIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I9M
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,4-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,4-DIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I90
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-8520 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 4-AMINO-3,4-DIHYDRO-2-(3-METHOXYPROPYL)-, 1,1-DIOXIDE, (R) | | Descriptor: | 4-AMINO-6-[N-(3-METHOXYLPROPYL)-2H-THIENO[3,2-E][1,2]THIAZINE 1,1-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9O
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,3,4-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I91
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6619 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-HYDROXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-HYDROXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9P
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,4,6-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(2,4,6-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I9L
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(4-FLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(4-FLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1I8Z
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH AL-6629 2H-THIENO[3,2-E]-1,2-THIAZINE-6-SULFONAMIDE, 2-(3-METHOXYPHENYL)-3-(4-MORPHOLINYL)-, 1,1-DIOXIDE | | Descriptor: | 6-[N-(3-METHOXY-PHENYL)-3-(MORPHOLIN-4-YLMETHYL)-2H-THIENO[3,2-E]-1,2-THIAZINE-1,1,-DIOXIDE]-SULFONAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chang, J.S, Liao, J, May, J.A, Christianson, D.W. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural aspects of isozyme selectivity in the binding of inhibitors to carbonic anhydrases II and IV.
J.Med.Chem., 45, 2002
|
|
1I9Q
 
 | | CARBONIC ANHYDRASE II (F131V) COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | 4-(AMINOSULFONYL)-N-[(3,4,5-TRIFLUOROPHENYL)METHYL]-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Kim, C.-Y, Chandra, P.P, Jain, A, Christianson, D.W. | | Deposit date: | 2001-03-20 | | Release date: | 2001-03-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fluoroaromatic-fluoroaromatic interactions between inhibitors bound in the crystal lattice of human carbonic anhydrase II.
J.Am.Chem.Soc., 123, 2001
|
|
1MQ7
 
 | | CRYSTAL STRUCTURE OF DUTPASE FROM MYCOBACTERIUM TUBERCULOSIS (RV2697C) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE | | Authors: | Sawaya, M.R, Chan, S, Segelke, B.W, Lekin, T, Heike, K, Cho, U.S, Naranjo, C, Perry, L.J, Yeates, T.O, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2002-09-13 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism.
J.Mol.Biol., 341, 2004
|
|
5KSV
 
 | | Crystal structure of HLA-DQ2.5-CLIP2 | | Descriptor: | HLA class II histocompatibility antigen gamma chain, MHC class II HLA-DQ-alpha chain, MHC class II HLA-DQ-beta-1 | | Authors: | Nguyen, T.B, Jayaraman, P, Bergseng, E, Madhusudhan, M.S, Kim, C.-Y, Sollid, L.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Unraveling the structural basis for the unusually rich association of human leukocyte antigen DQ2.5 with class-II-associated invariant chain peptides.
J. Biol. Chem., 292, 2017
|
|
5KSU
 
 | | Crystal structure of HLA-DQ2.5-CLIP1 at 2.73 resolution | | Descriptor: | HLA class II histocompatibility antigen gamma chain, HLA class II histocompatibility antigen, DQ alpha 1 chain, ... | | Authors: | Nguyen, T.-B, Jayaraman, P, Bergseng, E, Madhusudhan, M.S, Kim, C.-Y, Sollid, L.M. | | Deposit date: | 2016-07-10 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Unraveling the structural basis for the unusually rich association of human leukocyte antigen DQ2.5 with class-II-associated invariant chain peptides.
J. Biol. Chem., 292, 2017
|
|
7S6C
 
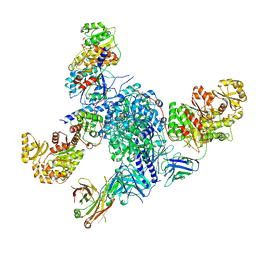 | | CryoEM structure of modular PKS holo-Lsd14 stalled at the condensation step and bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 4'-PHOSPHOPANTETHEINE, 6-deoxyerythronolide-B synthase EryA2, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
7S6D
 
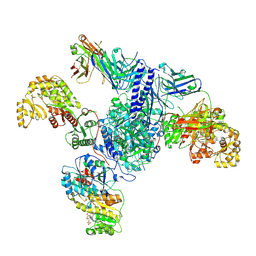 | | CryoEM structure of modular PKS holo-Lsd14 bound to antibody fragment 1B2, composite structure | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 6-deoxyerythronolide-B synthase EryA2, modules 3 and 4, ... | | Authors: | Bagde, S.R, Kim, C.-Y, Fromme, J.C. | | Deposit date: | 2021-09-13 | | Release date: | 2021-11-03 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Modular polyketide synthase contains two reaction chambers that operate asynchronously.
Science, 374, 2021
|
|
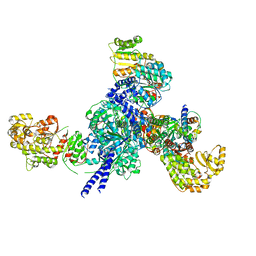
7S6B
 
 | |
7SH1
 
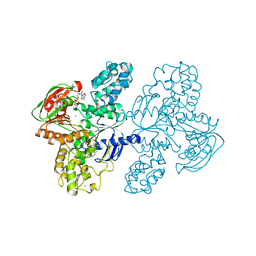 | | Class II UvrA protein - Ecm16 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Excinuclease ABC subunit UvrA, ... | | Authors: | Grade, P, Erlandson, A, Ullah, A, Mathews, I.I, Chen, X, Kim, C.-Y, Mera, P.E. | | Deposit date: | 2021-10-07 | | Release date: | 2022-10-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional analyses of the echinomycin resistance conferring protein Ecm16 from Streptomyces lasalocidi.
Sci Rep, 13, 2023
|
|
4RZM
 
 | | Crystal structure of the Lsd19-lasalocid A complex | | Descriptor: | CHLORIDE ION, Epoxide hydrolase LasB, FORMIC ACID, ... | | Authors: | Mathews, I.I, Hotta, K, Chen, X, Kim, C.-Y. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Epoxide hydrolase-lasalocid a structure provides mechanistic insight into polyether natural product biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
