1XCA
 
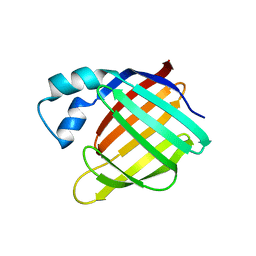 | | APO-CELLULAR RETINOIC ACID BINDING PROTEIN II | | Descriptor: | CELLULAR RETINOIC ACID BINDING PROTEIN TYPE II | | Authors: | Chen, X, Ji, X. | | Deposit date: | 1996-12-31 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of apo-cellular retinoic acid-binding protein type II (R111M) suggests a mechanism of ligand entry.
J.Mol.Biol., 278, 1998
|
|
4PZV
 
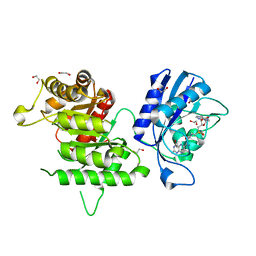 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with bisubstrate analog HPPK inhibitor J1D | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase/dihydropteroate synthase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structural enzymology and inhibition of the bi-functional folate pathway enzyme HPPK-DHPS from the biowarfare agent Francisella tularensis.
Febs J., 281, 2014
|
|
1YY7
 
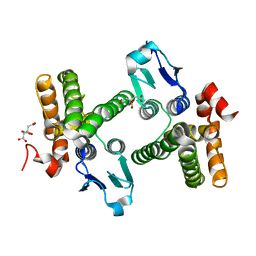 | | Crystal structure of stringent starvation protein A (SspA), an RNA polymerase-associated transcription factor | | Descriptor: | CITRIC ACID, stringent starvation protein A | | Authors: | Hansen, A.-M, Gu, Y, Li, M, Andrykovitch, M, Waugh, D.S, Jin, D.J, Ji, X. | | Deposit date: | 2005-02-23 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the function of stringent starvation protein A as a transcription factor
J.Biol.Chem., 280, 2005
|
|
1YYK
 
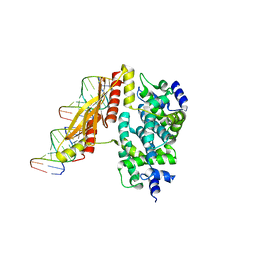 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYO
 
 | | Crystal structure of RNase III mutant E110K from Aquifex aeolicus complexed with double-stranded RNA at 2.9-Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YZ9
 
 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1YYW
 
 | | Crystal structure of RNase III from Aquifex aeolicus complexed with double stranded RNA at 2.8-Angstrom Resolution | | Descriptor: | 5'-R(*AP*AP*AP*UP*AP*UP*AP*UP*AP*UP*UP*U)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1ZYU
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with shikimate and amppcp at 2.85 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Shikimate kinase | | Authors: | Gan, J.H, Gu, Y.J, Li, Y, Yan, H.G, Ji, X. | | Deposit date: | 2005-06-11 | | Release date: | 2006-07-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue
Biochemistry, 45, 2006
|
|
2AJ3
 
 | | Crystal Structure of a Cross-Reactive HIV-1 Neutralizing CD4-Binding Site Antibody Fab m18 | | Descriptor: | Fab m18, Heavy Chain, Light Chain, ... | | Authors: | Prabakaran, P, Gan, J, Wu, Y.Q, Zhang, M.Y, Dimitrov, D.S, Ji, X. | | Deposit date: | 2005-08-01 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural mimicry of CD4 by a cross-reactive HIV-1 neutralizing antibody with CDR-H2 and H3 containing unique motifs.
J.Mol.Biol., 357, 2006
|
|
1ESR
 
 | |
1EX6
 
 | |
1EX7
 
 | |
1GBU
 
 | | DEOXY (BETA-(C93A,C112G)) HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vasquez, G.B, Ji, X, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1996-01-04 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cysteines beta93 and beta112 as probes of conformational and functional events at the human hemoglobin subunit interfaces.
Biophys.J., 76, 1999
|
|
1GBV
 
 | | (ALPHA-OXY, BETA-(C112G)DEOXY) T-STATE HUMAN HEMOGLOBIN | | Descriptor: | HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Vasquez, G.B, Ji, X, Pechik, I, Fronticelli, C, Gilliland, G.L. | | Deposit date: | 1995-12-20 | | Release date: | 1997-01-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cysteines beta93 and beta112 as probes of conformational and functional events at the human hemoglobin subunit interfaces.
Biophys.J., 76, 1999
|
|
7KDR
 
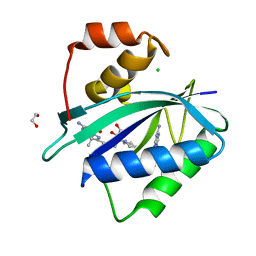 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-75 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]sulfonyl}-5'-deoxyadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.488 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
7KDO
 
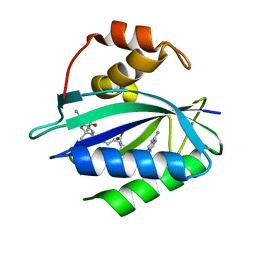 | | Crystal structure of Escherichia coli HPPK in complex with bisubstrate inhibitor HP-73 | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G.X, Shi, G, Ji, X. | | Deposit date: | 2020-10-09 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bisubstrate inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: Transition state analogs for high affinity binding.
Bioorg.Med.Chem., 29, 2021
|
|
3UDE
 
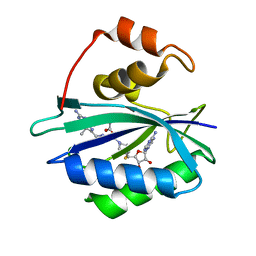 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1B | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UD5
 
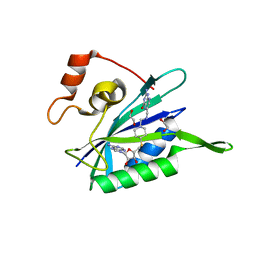 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1A | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
3UDV
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1C | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[1-(2-{[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]amino}ethyl)piperidin-4-yl]-5'-thioadenosine, ACETATE ION | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2011-10-28 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Bisubstrate analogue inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New design with improved properties.
Bioorg.Med.Chem., 20, 2012
|
|
2EZ6
 
 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
4F7V
 
 | | Crystal structure of E. coli HPPK in complex with bisubstrate analogue inhibitor J1D (HP26) | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-{[2-({N-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridin-6-yl)carbonyl]glycyl}amino)ethyl]sulfonyl}-5'-deoxyadenosine | | Authors: | Shaw, G, Shi, G, Ji, X. | | Deposit date: | 2012-05-16 | | Release date: | 2012-07-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Bisubstrate analog inhibitors of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase: New lead exhibits a distinct binding mode.
Bioorg.Med.Chem., 20, 2012
|
|
5T16
 
 | |
2G1K
 
 | | Crystal structure of Mycobacterium tuberculosis shikimate kinase in complex with shikimate at 1.75 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
2G1J
 
 | | Crystal structure of Mycobacterium tuberculosis Shikimate Kinase at 2.0 angstrom resolution | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Gan, J, Gu, Y, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2006-02-14 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Mycobacterium tuberculosis Shikimate Kinase in Complex with Shikimic Acid and an ATP Analogue.
Biochemistry, 45, 2006
|
|
2HK9
 
 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with shikimate and NADP+ at 2.2 angstrom resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
