5TFC
 
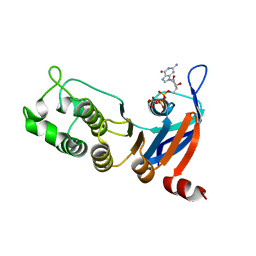 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with GTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFD
 
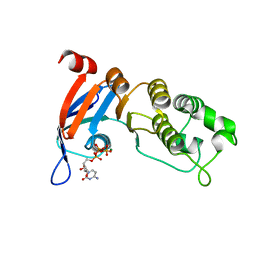 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFB
 
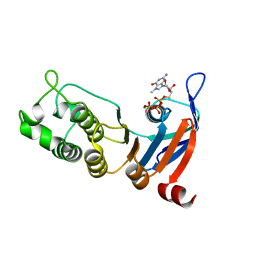 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with 7-methyl-GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFI
 
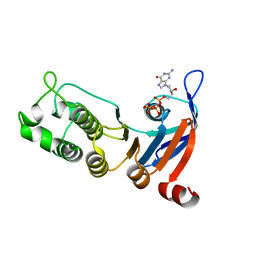 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFA
 
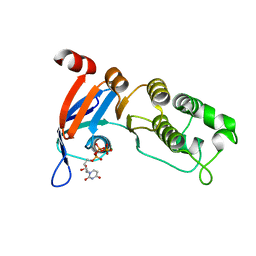 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dUTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, DEOXYURIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFG
 
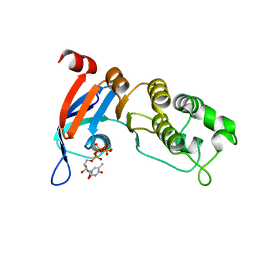 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with 5-methyl-UTP | | Descriptor: | 5-Methyluridine triphosphate, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
4DRT
 
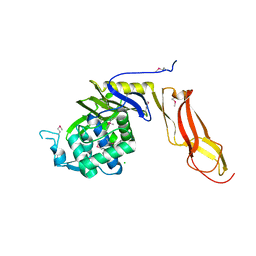 | | Three dimensional structure of de novo designed serine hydrolase OSH26, Northeast Structural Genomics Consortium (NESG) target OR89 | | Descriptor: | CHLORIDE ION, SODIUM ION, de novo designed serine hydrolase, ... | | Authors: | Kuzin, A, Su, M, Rajagopalan, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4DMB
 
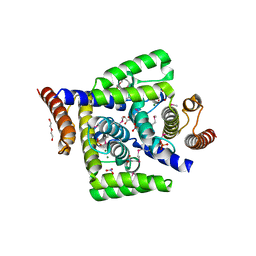 | | X-ray structure of human hepatitus C virus NS5A-transactivated protein 2 at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HR6723 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HD domain-containing protein 2, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-02-07 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR6723
To be Published
|
|
4DIU
 
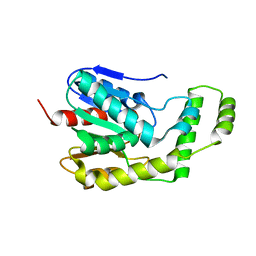 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94 | | Descriptor: | Engineered Protein PF00326 | | Authors: | Seetharaman, J, Lew, S, Wang, D, Kohan, E, Patel, D, Whitehead, T, Fleishman, S, Ciccosanti, C, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94
To be Published
|
|
4DLH
 
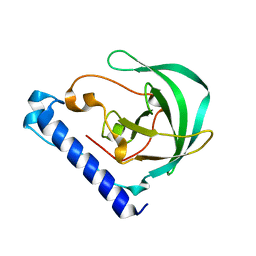 | | Crystal Structure of the protein Q9HRE7 from Halobacterium salinarium at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HsR50 | | Descriptor: | uncharacterized protein | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-02-06 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HsR50
To be Published
|
|
4E0A
 
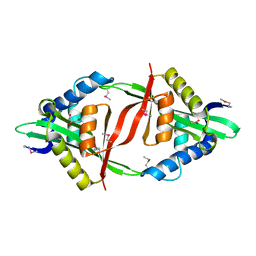 | | Crystal Structure of the mutant F44R BH1408 protein from Bacillus halodurans, Northeast Structural Genomics Consortium (NESG) Target BhR182 | | Descriptor: | ACETIC ACID, BH1408 protein, GLYCEROL, ... | | Authors: | Kuzin, A, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-02 | | Release date: | 2012-05-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Northeast Structural Genomics Consortium Target BhR182
To be Published
|
|
4E88
 
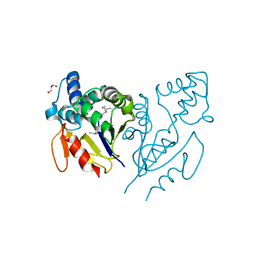 | | CRYSTAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, GLYCEROL | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Xiao, X, Sahdev, S, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-03-19 | | Release date: | 2012-04-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
4EVW
 
 | | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9. Northeast Structural Genomics Consortium (NESG) Target VcR193. | | Descriptor: | MAGNESIUM ION, Nucleoside-diphosphate-sugar pyrophosphorylase | | Authors: | Vorobiev, S, Neely, H, Su, M, Seetharaman, J, Mao, M, Xiao, R, Kohan, E, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the nucleoside-diphosphate-sugar pyrophosphorylase from Vibrio cholerae RC9.
To be Published
|
|
4ESS
 
 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ETJ
 
 | | Crystal Structure of E6H variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR185 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, CHLORIDE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-24 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4ZHN
 
 | | Crystal Structure of AlkB T208A mutant protein in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, COBALT (II) ION, ... | | Authors: | Ergel, B, Yu, B, Vorobiev, S.M, Forouhar, F, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | DFT Studies of the Rate-Limiting Step in the Reaction Cycle of the Fe/2OG Dioxygenase AlkB and Related Experimental Studies
To be published
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
6E12
 
 | | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141 | | Descriptor: | Alr8543 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Xiao, R, Ciccosanti, C, Patel, D, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141
To Be Published
|
|
1IY9
 
 | | Crystal structure of spermidine synthase | | Descriptor: | Spermidine synthase | | Authors: | Tan, A.Y, Smith, P.C, Shen, J, Xiao, R, Acton, T, Rost, B, Montelione, G, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-07-26 | | Release date: | 2002-10-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of spermidine synthase
To be Published
|
|
1JOD
 
 | |
1JOB
 
 | |
1KXZ
 
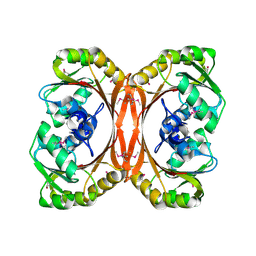 | | MT0146, the Precorrin-6y methyltransferase (CbiT) homolog from M. Thermoautotrophicum, P1 spacegroup | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, DeTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-01 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of MT0146/CbiT Suggests that the Putative
Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L3I
 
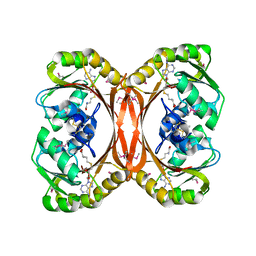 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, ADOHCY BINARY COMPLEX | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-27 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Crystal Structure of Mt0146/CbiT Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
1L2T
 
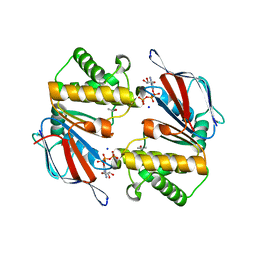 | | Dimeric Structure of MJ0796, a Bacterial ABC Transporter Cassette | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Hypothetical ABC transporter ATP-binding protein MJ0796, ISOPROPYL ALCOHOL, ... | | Authors: | Smith, P.C, Karpowich, N, Rosen, J, Hunt, J.F. | | Deposit date: | 2002-02-24 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer.
Mol.Cell, 10, 2002
|
|
1L3C
 
 | | MT0146, THE PRECORRIN-6Y METHYLTRANSFERASE (CBIT) HOMOLOG FROM M. THERMOAUTOTROPHICUM, C2 SPACEGROUP WITH SHORT CELL | | Descriptor: | Precorrin-6y methyltransferase/putative decarboxylase | | Authors: | Keller, J.P, Smith, P.M, Benach, J, Christendat, D, deTitta, G, Hunt, J.F. | | Deposit date: | 2002-02-26 | | Release date: | 2002-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Crystal Structure of Mt0146/Cbit Suggests that the Putative Precorrin-8W Decarboxylase is a Methyltransferase
Structure, 10, 2002
|
|
