8GPE
 
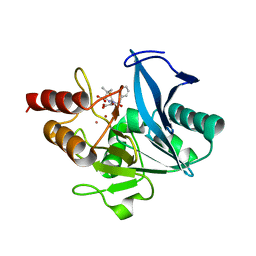 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed penicillin G | | 分子名称: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, POTASSIUM ION, ... | | 著者 | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | 登録日 | 2022-08-26 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
8GPC
 
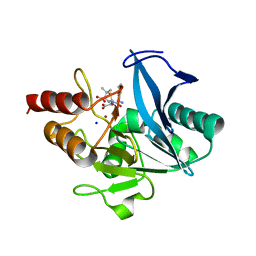 | | Crystal structure of NDM-1 at pH5.5 (Succinate) in complex with hydrolyzed ampicillin | | 分子名称: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo beta lactamase NDM-1, SODIUM ION, ... | | 著者 | Shi, X, Dai, Y, Zhang, Q, Liu, W. | | 登録日 | 2022-08-26 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Interplay between the beta-lactam side chain and an active-site mobile loop of NDM-1 in penicillin hydrolysis as a potential target for mechanism-based inhibitor design.
Int.J.Biol.Macromol., 262, 2024
|
|
5Z78
 
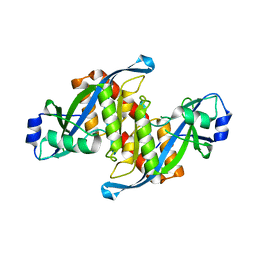 | | Structure of TIRR/53BP1 complex | | 分子名称: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | 著者 | Dai, Y.X, Shan, S. | | 登録日 | 2018-01-27 | | 公開日 | 2018-06-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.762 Å) | | 主引用文献 | Structural basis for recognition of 53BP1 tandem Tudor domain by TIRR
Nat Commun, 9, 2018
|
|
4N70
 
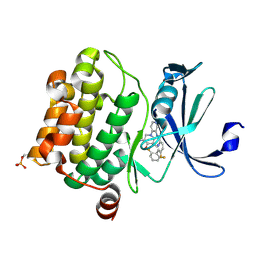 | | Pim1 Complexed with a pyridylcarboxamide | | 分子名称: | N-{4-[(3R,4R,5S)-3-amino-4-hydroxy-5-methylpiperidin-1-yl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | 登録日 | 2013-10-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4N6Y
 
 | | Pim1 Complexed with a phenylcarboxamide | | 分子名称: | 2-(acetylamino)-N-[2-(piperidin-1-yl)phenyl]-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | 登録日 | 2013-10-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
5DWR
 
 | | Identification of N-(4-((1R,3S,5S)-3-amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1,2 and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies | | 分子名称: | N-{4-[(1R,3S,5S)-3-amino-5-methylcyclohexyl]pyridin-3-yl}-6-(2,6-difluorophenyl)-5-fluoropyridine-2-carboxamide, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C, Bussiere, D, Burger, M. | | 登録日 | 2015-09-22 | | 公開日 | 2015-11-11 | | 最終更新日 | 2015-11-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies.
J.Med.Chem., 58, 2015
|
|
4N6Z
 
 | | Pim1 Complexed with a pyridylcarboxamide | | 分子名称: | 3-amino-N-{4-[(3S)-3-aminopiperidin-1-yl]pyridin-3-yl}pyrazine-2-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C.R, Le, V, Shu, W, Burger, M.T, Bussiere, D. | | 登録日 | 2013-10-14 | | 公開日 | 2013-11-06 | | 最終更新日 | 2014-07-02 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure Guided Optimization, in Vitro Activity, and in Vivo Activity of Pan-PIM Kinase Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4ZSA
 
 | | Crystal structure of FGFR1 kinase domain in complex with 7n | | 分子名称: | 4-(4-ethylpiperazin-1-yl)-N-[6-(3-methoxyphenyl)-2H-indazol-3-yl]benzamide, Fibroblast growth factor receptor 1 | | 著者 | Liu, Q.F, Xu, Y.C. | | 登録日 | 2015-05-13 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design, synthesis and biological evaluation of novel FGFR inhibitors bearing an indazole scaffold.
Org.Biomol.Chem., 13, 2015
|
|
6IUO
 
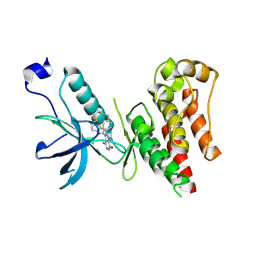 | | Crystal structure of FGFR4 kinase domain in complex with a covalent inhibitor | | 分子名称: | Fibroblast growth factor receptor 4, N-({4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}methyl)prop-2-enamide | | 著者 | Xu, Y, Liu, Q. | | 登録日 | 2018-11-29 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6ITJ
 
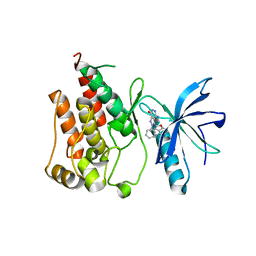 | | Crystal structure of FGFR1 kinase domain in complex with compound 3 | | 分子名称: | 4-azanyl-3-(3,5-dimethyl-1-benzofuran-2-yl)-2-phenyl-6~{H}-pyrazolo[3,4-d]pyridazin-7-one, Fibroblast growth factor receptor 1 | | 著者 | Xu, Y, Liu, Q. | | 登録日 | 2018-11-23 | | 公開日 | 2019-10-23 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.994 Å) | | 主引用文献 | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6IUP
 
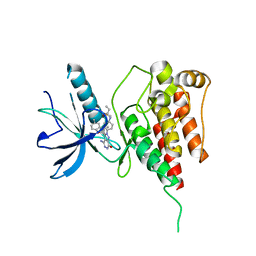 | | Crystal structure of FGFR4 kinase domain in complex with compound 5 | | 分子名称: | DIMETHYL SULFOXIDE, Fibroblast growth factor receptor 4, N-{4-[4-amino-3-(3,5-dimethyl-1-benzofuran-2-yl)-7-oxo-6,7-dihydro-2H-pyrazolo[3,4-d]pyridazin-2-yl]phenyl}prop-2-enamide | | 著者 | Xu, Y, Liu, Q. | | 登録日 | 2018-11-29 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and Development of a Series of Pyrazolo[3,4-d]pyridazinone Compounds as the Novel Covalent Fibroblast Growth Factor Receptor Inhibitors by the Rational Drug Design.
J.Med.Chem., 62, 2019
|
|
6C7S
 
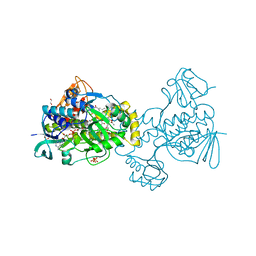 | | Structure of Rifampicin Monooxygenase with Product Bound | | 分子名称: | (1E,3S,4R,5S,6R,7R,8R,9S,10S,11E,13E)-15-amino-1-{[(2S)-5,7-dihydroxy-2,4-dimethyl-8-{(E)-[(4-methylpiperazin-1-yl)imino]methyl}-1,6,9-trioxo-1,2,6,9-tetrahydronaphtho[2,1-b]furan-2-yl]oxy}-7,9-dihydroxy-3-methoxy-4,6,8,10,14-pentamethyl-15-oxopentadeca-1,11,13-trien-5-yl acetate, 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Liu, L.-K, Tanner, J.J. | | 登録日 | 2018-01-23 | | 公開日 | 2018-04-18 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Evidence for Rifampicin Monooxygenase Inactivating Rifampicin by Cleaving Its Ansa-Bridge.
Biochemistry, 57, 2018
|
|
8G92
 
 | | Structure of inhibitor 16d-bound SPNS2 | | 分子名称: | 3-[3-(4-decylphenyl)-1,2,4-oxadiazol-5-yl]propan-1-amine, Sphingosine-1-phosphate transporter SPNS2 | | 著者 | Chen, H, Li, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-05-24 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Structural and functional insights into Spns2-mediated transport of sphingosine-1-phosphate.
Cell, 186, 2023
|
|
5HHF
 
 | |
5IIS
 
 | | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl-amide scaffold | | 分子名称: | 3-amino-N-(2'-amino-6'-methyl[4,4'-bipyridin]-3-yl)-6-(2-fluorophenyl)pyridine-2-carboxamide, DI(HYDROXYETHYL)ETHER, Serine/threonine-protein kinase pim-1 | | 著者 | Bellamacina, C, Bussiere, D, Burger, M. | | 登録日 | 2016-03-01 | | 公開日 | 2016-04-06 | | 最終更新日 | 2016-05-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Design, synthesis and structure activity relationship of potent pan-PIM kinase inhibitors derived from the pyridyl carboxamide scaffold.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5CPR
 
 | | The novel SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity | | 分子名称: | 6,7-dichloro-N-cyclopentyl-4-(pyridin-4-yl)phthalazin-1-amine, Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ... | | 著者 | Jakob, C.G, Upadhyay, A.K, Sun, C. | | 登録日 | 2015-07-21 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity.
Nat. Chem. Biol., 13, 2017
|
|
5Z0S
 
 | |
6JQX
 
 | |
6JQW
 
 | |
8GYA
 
 | |
8GY9
 
 | |
8GYB
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | 分子名称: | Methyltransferase | | 著者 | Chen, H, Lin, S, Lu, G.W. | | 登録日 | 2022-09-21 | | 公開日 | 2023-09-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8HR2
 
 | |
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | 分子名称: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | 著者 | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | 登録日 | 2021-08-24 | | 公開日 | 2022-08-31 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
